Hi everyone!
I have been working with the projpred package lately and I don’t feel very confident with the results. I started using stepwise regression when I found this package, and I think is the tool I need. I have 39 variables/features (x1,x2,x3,…,x39) and I want to reproduce the response variable y. My prior knowledge is that some of the variables may be not relationated with y, and some of them are highly correlationated with the others, so the information the carry is redundant. The idea is then to find a minimal subset of the variables to reproduce the y value.
I tried with the horseshoe prior as indicated by Piironen and Vehtari (2017):
formula <- y ~ x1 + x2 + x3 + x4 + x5 + x6 + x7 + x8 + x9 + x10 + x11 + x12 + x13 + x14 + x15 + x16 + x17 + x18 + x19 + x20 + x21 + x22 + x23 + x24 + x25 + x26 + x27 + x28 + x29 + x30 + x31 + x32 + x33 + x34 + x35 + x36 + x37 + x38 + x39
n <- nrow(data)
D <- 39 # number of parameters
p0 <- 15 # prior guess for the number of relevant variables
tau0 <- p0/(D-p0) * 1/sqrt(n)
fit <- brm(formula, family=gaussian(), data=data,
prior=prior(horseshoe(scale_global = tau0, scale_slab = 1), class=b),
seed=123456789, chains=4, iter=2000, warmup=1000,
control=list(max_treedepth=12,adapt_delta=0.99), save_pars=save_pars(all=TRUE))
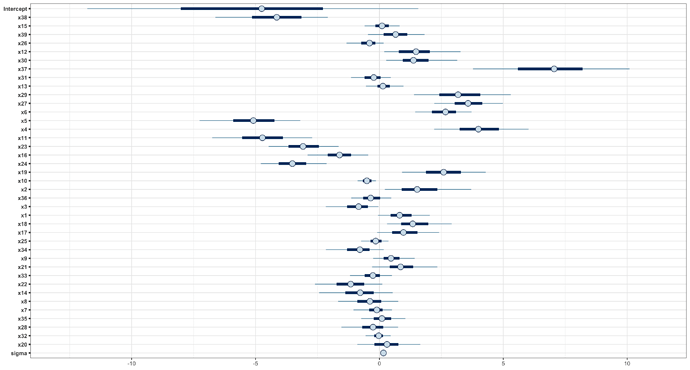
The results, with mcmc_intervals, are:
(the order of the variables is the one got by cv_varsel, see later) Note the high dispersion/error on the Intercept. This is a problem I think, since the range of my response variable is range(y) = [-1.58 , 0.37], so I don’t know if I can be confident about the predictions for y with that uncertainty.
Let’s go to proper projpred. I construct the reference model and use the cv_varsel function:
refmodel <- get_refmodel(fit)
vs <- cv_varsel(refmodel, method = "forward", nterms_max = 50)
plot(vs, stats = c('elpd', 'rmse'))

I get this plot, and using the suggest_size function with ‘elpd’ or ‘rmse’, it suggests me to use 19 variables. So, now:
proj <- project(vs, nterms = 19, ns = 500)
mcmc_intervals(as.matrix(proj))
The Intercept has a large dispersion again, and some of the coefficients (x31, x13) are nearly zero! I don’t feel very confident about this.
Finally, I plot the predictions using the reference model (blue x) and the submodel (black dots) versus the original y values, with the 1:1 relationship:

So it seems like projpred did a good job, reproducing with the submodel the predictions of the reference model. But I think it could be better, with less dispersion (i.e. the predictions closer to the 1:1 line), and better coefficients (that is, less dispersion for the intercept, coeffs nearly zero?, etc.)
Does someone know if I can do better? May be a problem with the reference model?