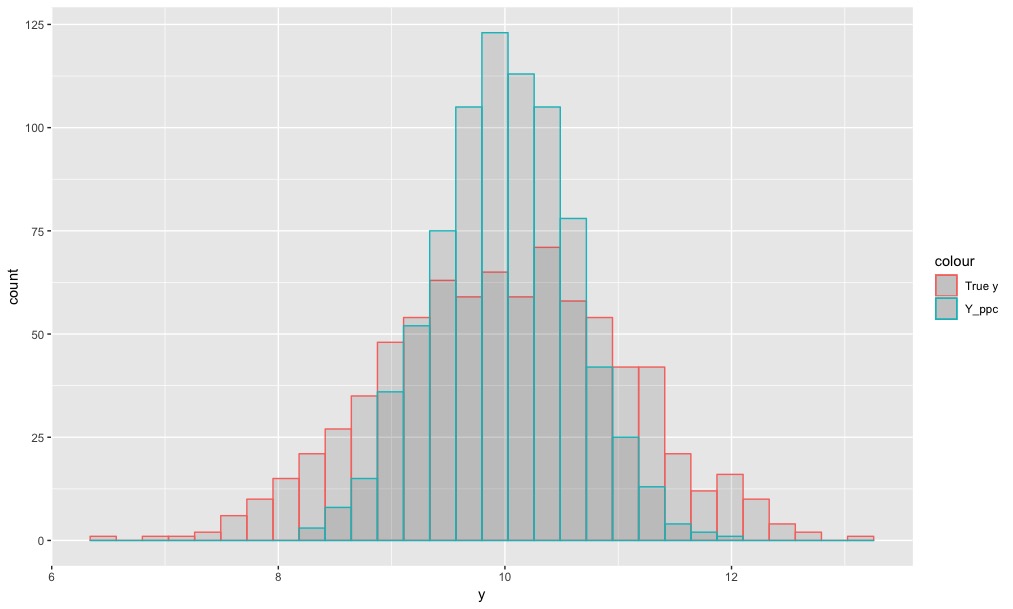
So I have a multi-normal model that behaves pretty well but when I do a posterior predictive check the y_ppc is a bit too confined when compared to the data Y value:
I thought I could change this by changing from a multi-normal to a multi_student_t distirbution.I set nu =2 rather than a parameter as I thought this would be an easier fit. However running this model gives me initialisation errors: Initialization between (-2, 2) failed after 100 attempts. z before crashing. I set init = 0 and then it will run but I’m getting almost all divergences.
Can any advise on where I’m going wrong here? I thought I was making a small change to minize issues but it seems not! Code below:
data{
int<lower=0> NvarsX; // num independent variables
int<lower=0> NvarsY; // num dependent variables
int<lower=0> N; // Total number of rows
matrix[N, NvarsX] x; // data for independent vars
vector[NvarsY] y [N]; // data for dependent vars
}
parameters{
cholesky_factor_corr[NvarsY] L_Omega; //For the correlations between outcome variables
vector<lower=0>[NvarsY] L_sigma; //Residual SD of outcome variables
vector[NvarsY] Beta0; // Intercepts for each Y
vector[NvarsX] Beta [NvarsY]; // Coefficients for each X vs each Y
}
transformed parameters{
vector[NvarsY] mu[N];
matrix[NvarsY, NvarsY] L_Sigma;
L_Sigma = diag_pre_multiply(L_sigma, L_Omega);
for (i in 1:N){
for (dv in 1:NvarsY){
mu[i, dv] = Beta0[dv] + dot_product(to_vector(Beta[dv, 1:NvarsX]), x[i, 1:NvarsX]);
}
}
}
model{
//Priors
L_Omega ~ lkj_corr_cholesky(1);
L_sigma ~ normal(0, 2.5);
Beta0 ~ normal(0, 3);
for (dv in 1:NvarsY){
Beta[dv, 1:NvarsX] ~ normal(0, 2);
}
//likelihood
//y ~ multi_normal_cholesky(mu, L_Sigma);
y ~ multi_student_t(2, mu, L_Sigma);
}
generated quantities{
matrix[NvarsY, NvarsY] Omega;
matrix[NvarsY, NvarsY] Sigma;
vector[NvarsY] mu_ppc[N]; // model predictions for posterior predictive check
vector[NvarsY] y_ppc [N]; // model predictions for posterior predictive check
// Generate correlation matrix
Omega = multiply_lower_tri_self_transpose(L_Omega);
Sigma = quad_form_diag(L_Omega, L_sigma);
// Generate y_ppc
for (n in 1:N){
for(dv in 1:NvarsY){
mu_ppc[n, dv] = Beta0[dv] + dot_product(to_vector(Beta[dv, 1:NvarsX]), x[n, 1:NvarsX]);
}
}
//y_ppc = multi_normal_cholesky_rng(mu_ppc, L_Sigma);
y_ppc = multi_student_t_rng(2, mu_ppc, L_Sigma);
}