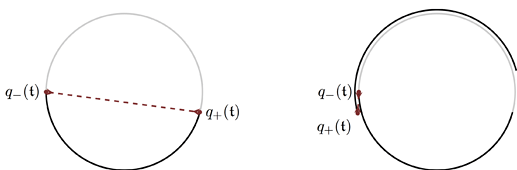This is most likely related to the performance regression discussed by @bbbales2 and @betanalpha in the thread on stepsize adaptation tweak but not directly relevant for that thread’s topic.
NUTS is supposed to automatically select the optimal integration time for HMC and because that should only depend on the geometry of the model I would expect that the number of leapfrog steps NUTS uses for a given model is inversely proportional to the stepsize. That is an overly simplistic argument but even so I was quite surprised by the results of some simulations.
All fits here were done using PyStan 2.19.
The model I tested is a five dimensional vector whose all components have IID standard normal distribution. To keep things simple I used the unit_e metric (which is optimal for standard normal anyway), no warmup, four chains, run for 1000 iterations per chain. Here’s the results of fitting it with 100 different stepsizes:

No divergent transitions occurred during the runs. Even to the right of the vertical red line (where the integrator is theoretically unstable) NUTS criterion cut off the trajectories before they were diagnosed as divergent.
The top graph shows the effective sample sizes for lp__ and the five components of the vector x and grey line is the nominal sample size of 4000. The middle graph shows the average accept_stat__ over the iterations and the bottom graph shows the average n_leapfrog__. Note the logarithmic scale, those spikes are huge.
The problem can be present during warmup even if adaptation eventually finds a stepsize with stable treedepth. Here’s a 500 iteration warmup run for a model of 400 IID standard normals. All control parameters except warmup duration are left to their default values.

When we plot the n_leapfrog__ of each iteration against its stepsize a pattern emerges and it corroborates what can be seen in the first graph.

The vertical grey line is where stepsize freezes after adaptation. Luckily it is in a region of stable treedepth and the model fitting proceeds smoothly with 15 leapfrog steps per posterior draw. Models with 100 to 300 parameters aren’t always as lucky.
Anyway, it seems that whenever the sampler needs to change the treedepth it must visit maximum treedepth first. Let’s think about why that happens.
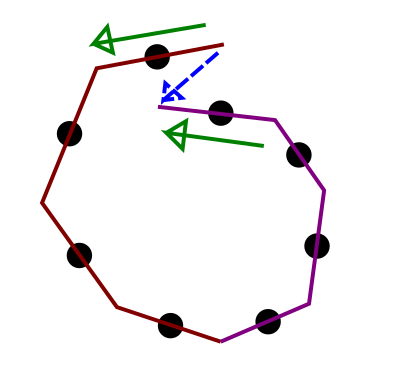
The sampler builds a trajectory as a binary tree, repeatedly concatenating subtrees until it finds a tree that makes a U-turn. Since we’re sampling from a standard normal distribution the final trajectory will typically look something like three quarters of a circle/ellipse around the origin.
As the stepsize decreases, integration time shrinks proportionally until the trajectory becomes shorter than a semicircle so that it doesn’t quite make a U-turn anymore and the sampler needs to double the number of steps. But two 180 degree U-turns add up to a full 360 degree turn which looks a lot like no turn at all. Therefore at the precise stepsize where treedepth should increase by one, U-turns become invisible and NUTS does not stop until it hits max_treedepth. When stepsize is decreased further the circular trajectory shortens enough to clearly be a steep U-turn and NUTS stabilizes at a new treedepth. This explains what we see in the graphs above.
But hold on, isn’t that impossible? Each subtree is slightly less than a U-turn so their concatenation must be slightly less than a full circle, and that looks like a backward step which the NUTS criterion will detect and stop at. Now, that would indeed be the case if the first point of the second subtree were also the last point on the first subtree but there is in fact one leapfrog step between them. That additional kink in the middle allows two almost-semicircles to overshoot a full circle and avoid termination by NUTS.
A simple fix might be to check the U-turn condition not just at the ends but also in the middle of the new tree. This can be done by changing compute_criterion() in base_nuts.hpp to
virtual bool compute_criterion(Eigen::VectorXd& p_sharp_minus,
Eigen::VectorXd& p_sharp_middle,
Eigen::VectorXd& p_sharp_plus,
Eigen::VectorXd& rho) {
return p_sharp_plus.dot(rho) > 0
&& p_sharp_middle.dot(rho) > 0
&& p_sharp_minus.dot(rho) > 0;
}
Here p_sharp_middle is the sum of p_sharp at the end of the left subtree and at the start of the right subtree.
Here’s the first graph again after but now using the modified NUTS criterion.