I’ve been working for a while on a template for Python-based Bayesian workflow projects. I discussed a previous version here and presented a more up to date one at the last Stancon, but there have been a few more changes and I’m very keen to find out what the Stan community thinks, so I thought I’d share again!
The target audience for bibat are people who want to implement a Bayesian statistical analysis project using Python, Stan and arviz. More specifically, bibat targets analyses that might involve many interacting components like models, datasets and ways of fitting a model to a dataset.
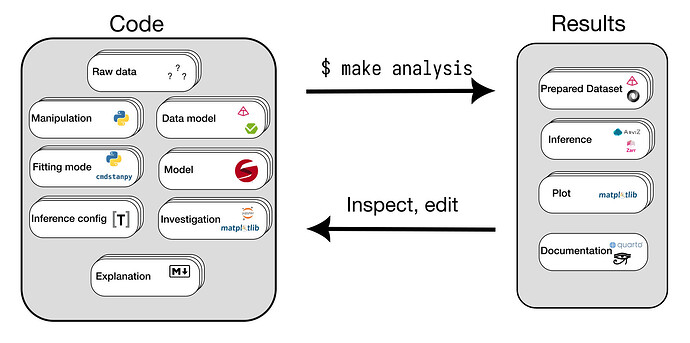
The idea is to implement a workflow that looks like this:
For a specific example of what a project made using this workflow looks like, see here.
I think this workflow is nice for Bayesian analysis projects because
- There is a a single command that runs everything, making the analysis easy to replicate.
- Most components of the analysis are modular and potentially plural, so the analysis scales gracefully.
- It includes data validation and documentation, so it is more likely that the analysis is correct and that plots in the paper are up to date.
With bibat you can implement this workflow by following these steps:
> pipx install copier> copier copy gh:teddygroves/bibat my_analysis- answer some questions about your analysis in the terminal
> cd my_analysis> make analysis (nb at this stage bibat may attempt to install cmdstan on your computer)
Now you’re in the workflow loop!
To find out more about bibat, check out the documentation, source code and particularly this vignette that describes in detail how to go from these steps to the example project linked above.
Please let me know what you think, especially about these questions:
- Is bibat general enough to cover a sufficiently wide range of use cases? Is something big that can’t currently be done?
- Is the template project simple enough that you can easily understand what each part does and how you would change it?
Both of these points came up in the previous discussion and I’ve done a lot of work towards addressing them, but it’s difficult to accurately judge so outside feedback would be very much appreciated!
8 Likes
Thanks for posting, @Teddy_Groves1. This looks really interesting. And I’ve moved over to Python full time, so I’m happy this is using cmdstanpy.
I think we’re all struggling with the problems this addresses. For example, I’m currently evaluating 18 IRT-like crowdsourcing models, each of which is defined by removing a subset of 5 features from a full model (14 models were dupes once identified). It was a bit of a pain to get the code arranged so that I didn’t have a ton of duplication (4 includes per file do the heavy lifting), but I’m imagining it would have been a lot harder for someone less familiar with Stan than me. But now I’m faced with having to analyze all the outputs. It’s so painful I started another project and plan to come back to this one next week. One issue I have is that each of the 5 features is meaningful and I sort of want to report on them in groups (like the group that separates sensitivity/specificity from the one that doesn’t).
P.S. I was checking out your mRNA example and it looks like a lot of extraneous compute-generated files were checked in accidentally like lognormal and lognormal.hpp.
P.P.S. Why is bibat creating a code of conduct for every project? There’s a danger those will conflict with a project’s existing code of conduct when imported and might cause confusion if auto-generated and then put up on a web site somewhere.
1 Like
Thanks a lot for the interest and feedback!
I haven’t thought much about the problem of avoiding repeating Stan code (though I did deliberately add an include to the starter model to highlight that they exist).
The problem with keeping track of which models have which features seems annoying, and is also something that bibat currently doesn’t help with! If you are using arviz to store your models’ outputs (and if I understood the problem description correctly), it might help to know that InferenceData objects come with a dictionary called attrs where you can store arbitrary metadata. I think it would be possible to label your models according to their features by doing something like this:
import arviz as az
idata = az.load_arviz_data("rugby")
idata.attrs["is_about_rugby"] = True
Thanks! These are now deleted. That example is a little out of date now - the baseball vignette and example project is the only one I’m keeping fully up to date.
This is a good point. I included the code of conduct in the template project because I thought that it would make it more likely for people to use a code of conduct, plus templating makes it easy to quickly create a custom one, which can save some time. However a) as you say there are cases where someone wants to use the template project but they already have a code of conduct, creating a risk of conflicts and b) it is maybe a bit prescriptive to assume that one choice is right for all projects that do want one.
Looking at some popular python templates I think there isn’t really a consensus about the best approach. cookiecutter-data-science and cookiecutter-django don’t put codes of conduct in their template projects, whereas cookiecutter-hypermodern-python and cookiecutter-cms provide a single option in basically the same way as bibat.
Overall I think the best approach is to add a choice to the questionnaire so that the user can at least choose not to add a code of conduct, and possibly also provide a list of different options for projects that do want one (I’ve added an issue for doing this).