Hello!
I was wondering if anyone had any experience with plotting predictions of a multivariate binomial model with brms? I will only provide the code for one response variable below.
All I get is this warning message:
Warning message:
Computation failed instat_eye()
Caused by error inbw.SJ():
! sample is too sparse to find TD
My_model <- brm(FutureReproduction~ Reproductive_success+ Age
+ Precipitation+ Scaled_Leg_index
+ Reproductive_success*Age
+ (1 | ID),
family = bernoulli())
condition <-expand.grid("Age" = (c(3, 4, 5, 6, 7,8, 9, 10, 11)),
Year = 2011,
ID = 3,
Precipitation= 0,
Leg_index= 0,
Reproductive_success = c(0, 1))
Model_prediction <- condition %>%
add_predicted_rvars(My_model, allow_new_levels = TRUE, re_formula = NA, newdata = .) %>%
mutate(Reproductive_success = factor(ifelse(Reproductive_success == 1, "Yes", "No"), levels = c("No", "Yes")))
> Model_prediction
# A tibble: 18 × 7
Age Year ID Precipitation Leg_index Reproductive_success prediction[,"Jumpheight"] [,"Sprintspeed"] [,"Future Reproduction"] [,"Survival"]
<dbl> <dbl> <dbl> <dbl> <dbl> <fct> <rvar[,1]> <rvar[,1]> <rvar[,1]> <rvar[,1]>
1 3 2011 3 0 0 No 21.33 ± 5.9 2.41 ± 1.8 0.261 ± 0.44 0.97 ± 0.18
2 4 2011 3 0 0 No 10.63 ± 6.0 1.57 ± 1.8 0.230 ± 0.42 0.97 ± 0.17
3 5 2011 3 0 0 No 4.60 ± 5.9 1.03 ± 1.8 0.202 ± 0.40 0.97 ± 0.18
4 6 2011 3 0 0 No 2.49 ± 5.9 0.74 ± 1.8 0.188 ± 0.39 0.96 ± 0.19
5 7 2011 3 0 0 No 2.63 ± 5.9 0.67 ± 1.8 0.168 ± 0.37 0.96 ± 0.19
6 8 2011 3 0 0 No 3.59 ± 5.9 0.64 ± 1.8 0.153 ± 0.36 0.95 ± 0.22
7 9 2011 3 0 0 No 5.12 ± 5.9 0.66 ± 1.8 0.148 ± 0.36 0.93 ± 0.25
8 10 2011 3 0 0 No 6.88 ± 6.0 0.69 ± 1.8 0.136 ± 0.34 0.90 ± 0.29
9 11 2011 3 0 0 No 8.97 ± 6.4 0.79 ± 1.9 0.134 ± 0.34 0.85 ± 0.35
10 3 2011 3 0 0 Yes 9.54 ± 6.6 -0.66 ± 2.0 0.029 ± 0.17 0.91 ± 0.28
11 4 2011 3 0 0 Yes 5.97 ± 5.9 -0.41 ± 1.8 0.025 ± 0.16 0.93 ± 0.25
12 5 2011 3 0 0 Yes 2.89 ± 6.1 -0.71 ± 1.8 0.022 ± 0.15 0.94 ± 0.23
13 6 2011 3 0 0 Yes 0.46 ± 6.0 -1.37 ± 1.8 0.018 ± 0.13 0.95 ± 0.23
14 7 2011 3 0 0 Yes -1.18 ± 6.0 -2.11 ± 1.8 0.016 ± 0.13 0.95 ± 0.23
15 8 2011 3 0 0 Yes -2.21 ± 6.1 -2.56 ± 1.8 0.017 ± 0.13 0.94 ± 0.24
16 9 2011 3 0 0 Yes -2.53 ± 6.1 -2.63 ± 1.8 0.018 ± 0.13 0.92 ± 0.27
17 10 2011 3 0 0 Yes -2.30 ± 6.2 -2.54 ± 1.8 0.017 ± 0.13 0.89 ± 0.32
18 11 2011 3 0 0 Yes -1.54 ± 6.4 -2.22 ± 1.9 0.019 ± 0.14 0.86 ± 0.35
#Plot
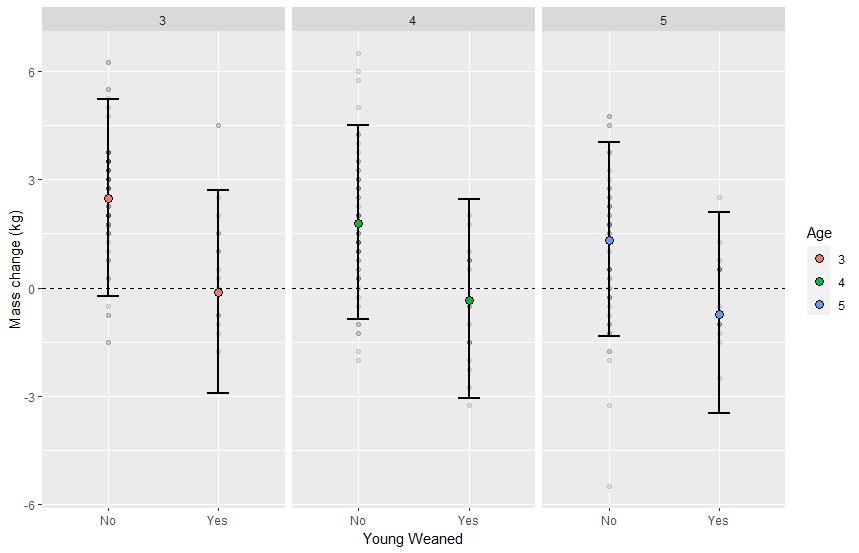
Model_prediction %>%
ggplot(aes(x = Reproductive_success , ydist = .prediction[,"Future Reproduction"]) )+
stat_eye()
Warning message:
Computation failed in `stat_eye()`
Caused by error in `bw.SJ()`:
! sample is too sparse to find TD
I used the exact same approach for my other figures with gaussian distribution (continuous response variables) and had no issues