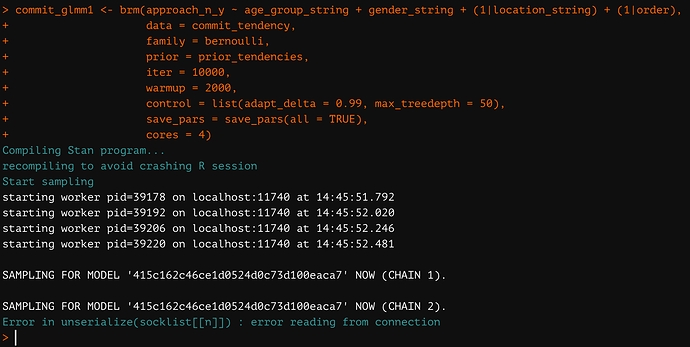
Hello,
I am having quite a lot of trouble using brms to fit a very simple mixed effects model on my desktop Mac (OS version: 10.14.6, R version: 4.1.1, brms version: 2.16.1). The model includes 2 fixed effects parameters and 2 random intercepts.
Specifically, I keep receiving the following error message: “Error in unserialize(socklist[[n]]) : error reading from connection”. I have attached a photo of the error as it most recently appeared in RStudio.
Some points about when the error message does and does not appear:
First, kindly note that in the attached photo I have “cores = 4.” If I use the default core number (=1), then RStudio simply crashes and no error message appears (also, e.g., Error in unserialize(socklist[[n]]) : error reading from connection). So, I know I can only use multiple cores to reliably obtain the error message.
Second, the error seems to appear only when I include the “control = list()” argument of the “brm” function. That is, the error does not appear if I run brm using the default values of the control parameters for the sampler; in this case, the sampler runs normally, without error. So, I know I can run my model if I exclude the “control = list()” argument.
Third, when using the same desktop computer the same error appears when I run a different dataset and script that includes a “control = list()” command. So, I know the issue is not somehow unique to the dataset or script I am working with.
Fourth, when using my personal laptop the error does not appear when I run the identical script (Mac OS version: 10.15.7, R version: 4.0.5, brms version: 2.16.1). So, I know the error is somehow arising from the specific desktop computer I am working with.
Some prior attempts at resolving this issue include:
I have tried uninstalling and reinstalling R, RStudio, brms and the various packages associated with it (e.g., rstan, rstanarm) and Xcode and deleted preexisting Makevars files (following Error in unserialize(socklist[[n]]) : error reading from connection Error in serialize(data, node$con, xdr = FALSE) : error writing to connection - #27 by bgoodri). In my most recent attempt at reinstalling rstan, I have followed the following instructions verbatim: RStan Getting Started · stan-dev/rstan Wiki · GitHub. However, none of these approaches has resolved the issue.
The best that I can discern is that there is some issue with converting the brms commands into C++ code. However, this may be wrong and, even if it is right, I have run out of ideas about how to proceed with resolving this issue.
Any help at all would be greatly appreciated, thank you.