Hi!
The discussions on parallel autodiff v2 lead to a few considerations
- we really would like to run the parallel phase to also run during the reverse sweep
- we concentrate on the reduce sum only => so only a single output
- we want to keep track of all var’s which are part of anything running parallel => this means we cannot use closures which have any var in them (@Bob_Carpenter I hope we can have a flag for the closures indicating if these capture data only)
With all these points we can actually code a map_rect style thing which only relies on our current nested AD. At least this is what I think can be done. However, the down-side is that quite a few vari’s need to be copied around and moreover to make this thing convenient to use we need to use C++ tuples to make use of the ... magic thing… and here I would very much appreciate help. More details below.
The best news is: This can be implemented on the back of our current AD system. No need to refactor the thing!
The only bad news is that this technique can likely not be used for higher-order autodiff, but oh well… if you need parallelism then you probably anyway cannot afford higher-order derivatives (which would anyway be better off being parallelized differently, I think).
What I have done is to code up a basic prototype with a restricted interface geared toward the example I use, a hierarchical poisson log-lik. For this case I did
- test that we get correct derivatives with/without threading…looks good
- test that nested parallelism works… it does
- write a benchmark to test if the extra copying work is harming us or not…its not so much harming (and can even be improved with more clever coding)
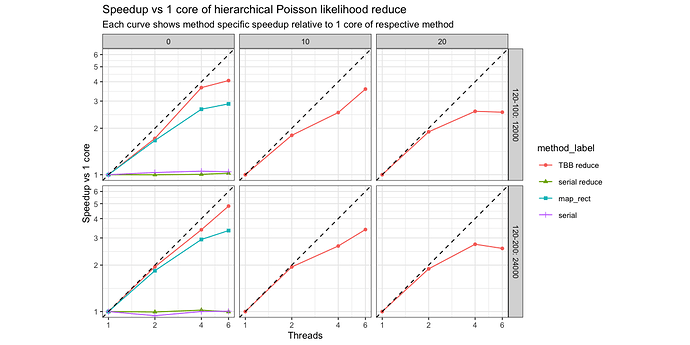
The benchmark results are for 12 \cdot 10^3 terms of a normalized log-likelihood which is grouped in 10 (dense) or 100 (sparse) groups. The dense case represents lots of data per group and this implies less copying overhead than the sparse case. I wanted to see how bad the copying overhead is hurting us… as there is not much of a difference we are fine from my perspective here.
The benchmarks are run with the perf-math repository on my 6-core MacBook Pro. So this is the profiled call to the grad call which should reflect the net speedup in an actual Stan model.
So we get quite decent speedups here which are totally usable. I should have the time next year to run the respective Stan model with this.
What is left?
I would really need a few hands here to help me out with the C++ magic which needs to happen to make this function a convenient function to use… and folks from the stanc3 compiler hopefully can also join to bind this to the Stan language.
The files are on the stan-math repo in the proto-parallel-v3 branch:
- prim version of
parallel_reduce_sum, here, with comments on where some help would be great - rev parts here
- unit tests which test gradients, nested parallelism and a hierarchical example, here
The benchmarks are from here. The repo contains, I think, all what is needed to get the above plots.
I am tagging a few people who I know are good with C++ magic for these tuples + type wrangling… @andrjohns @bbbales2 @tadej @stevebronder … sorry if I forgot you.
Should we drop the v2 parallel autodiff refactor thing? If all of what I did is correct… then probably the answer is yes for the moment being. Refactoring the AD sub system turns out to be a minefield. It takes a lot of energy from quite a few people to get this over, so it’s good if we can stay with what we have.
Happy new year!
Best,
Sebastian