I’m trying to develop a model that would be used for analyzing fMRI data. However, I’m running into parameter recovery issues. I would greatly appreciate some help or advice.
Background
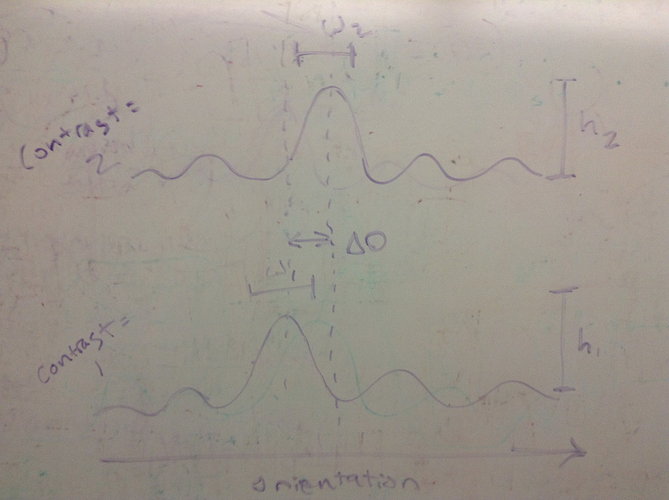
The overall goal is to explore the how well we can use fMRI to characterize the response of various orientation-selective channels of neurons. It is known that neurons have preferences for visual stimuli shown at particular orientations. Moreover, it is known how their response changes as the contrast of the stimuli change. So, among a number of different possible ways that contrast could affect the response of neurons, we’re trying to recover from fMRI data how their responses almost certainly changed (not at the level of individual neurons, but at least at the level of largish channels or groups of neurons).
Currently
At this point, I have what I thought was an okay model, but even when I use a very restricted form of the model I’m unable to recover the parameters that I used to generate the data.
Each observation, y, is distributed according to
y[v,c,o] ~ normal(mu[c,o], noise_voxel[v] )
where v is a voxel, c is the contrast, and o is the orientation being presented. noise_voxel is voxel-specific noise. μ[c,o] should represent the expected activity of a voxel shown a stimulus of a particular orientation at a given contrast, given a particular distribution of channels within the voxel. mu[c,o] would be distributed as a mixture of normal distributions – one for each K channels – corresponding to the proportion of each channel that is present in a voxel:
mu[c,o] = dot_product(channel_W, channel_resp[1:K,c,o])
such that
channel_resp[c,o,k] ~ normal(channel_mu[c,o,k], noise_channel[c] ).
The error, noise_channel, depends on the level of contrast (same error across all channels). w would be a simplex that is Dirichlet-distributed.
Finally, channel_mu is the expected response of a channel to a given orientation presented at a given contrast:
channel_mu[c,o,k] ~ M[c] * exp(von_mises_lpdf( testedTheta[o] | preferredOri[k], kappa[c] ));
a channel, k’s, response to an orientation, testedTheta[o], is a function of the scaled (by M) density of a von Mises – parameterized by the channel’s preferred orientation, preferredOri, as well as the contrast dependent parameters scale M, and concentration kappa.
I’ve restricted everything the model such that it’s just estimating kappa and channel_resp. However, the posterior of kappa is way off. For example, when I generate data with kappa = 1 (setting all other parameters to be data), the model returns a bi-modal posterior clustering around 0 and 0.2.
If it’s helpful, I’ve attached my attempt at coding this model. There is also an example of data that I’ve generated to test the code, and an R script to grabs the data and runs Stan. In that data (only 1 voxel), the true number of channels is locked to 2, there are 2 levels of contrast, 100 orientations were tested 100 times each, channel weights were set to 0.4 and 0.6, the preferred channel orientations were -pi/2 and pi/2, M was 5, and all noise was 0.1. As in the last paragraph says, the only parameter left to be estimated is kappa, whose true value is 1, but the posteriors cluster around 0 and 0.2.
vtf_restricted_wpm.stan (1.5 KB)
run_help.R (654 Bytes)
stan_data.R (446.8 KB)