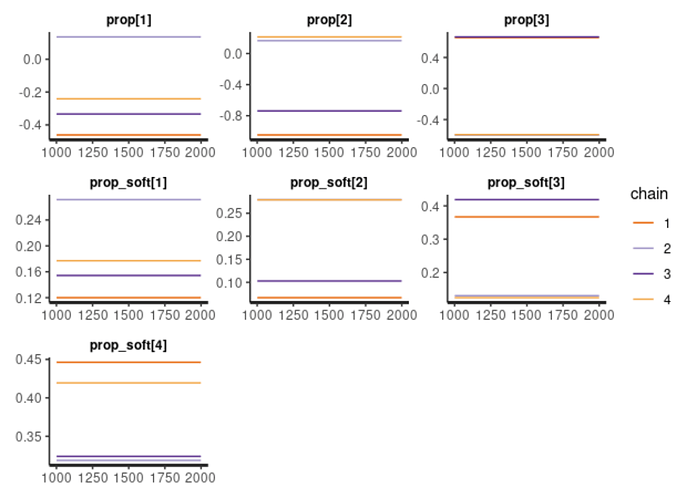
OK what is puzzling me that if I use this model with no jacobian, and with QR approach to N-1 degrees of freedom vector (Test: Soft vs Hard sum-to-zero constrain + choosing the right prior for soft constrain).
functions{
vector Q_sum_to_zero_QR(int N) {
vector [2*N] Q_r;
for(i in 1:N) {
Q_r[i] = -sqrt((N-i)/(N-i+1.0));
Q_r[i+N] = inv_sqrt((N-i) * (N-i+1));
}
return Q_r;
}
vector sum_to_zero_QR(vector x_raw, vector Q_r) {
int N = num_elements(x_raw) + 1;
vector [N] x;
real x_aux = 0;
for(i in 1:N-1){
x[i] = x_aux + x_raw[i] * Q_r[i];
x_aux = x_aux + x_raw[i] * Q_r[i+N];
}
x[N] = x_aux;
return x;
}
}
transformed data{
int K = 4;
vector[K] alpha = [1,2,3,4]';
matrix[K,K] ident = diag_matrix(rep_vector(1,K));
vector[2*K] Q_r = Q_sum_to_zero_QR(K);
real x_raw_sigma = inv_sqrt(1 - inv(K));
}
parameters{ vector[K-1] prop; }
transformed parameters {
simplex[K] prop_soft = softmax(sum_to_zero_QR(prop, Q_r));
}
model{
prop_soft ~ dirichlet(alpha);
prop ~ normal(0, x_raw_sigma);
}```
```r
fit2 = stan(model_code=stancode, cores=4, iter = 10000, warmup = 1000)
I get same marginal posterior of the simplex compared to the the source model
transformed data{ vector[4] alpha = [1,2,3,4]'; }
parameters{ simplex[4] prop; }
model{ prop ~ dirichlet(alpha);
fit= stan(model_code = stancode, cores=4, iter = 10000, warmup = 1000)
tidybayes::spread_draws(fit2, prop_soft[N]) %>%
select(N,prop_soft) %>%
arrange(N,prop_soft) %>%
mutate(i = 1:n()) %>%
left_join(
tidybayes::spread_draws(fit, prop[N]) %>%
select(N,prop) %>%
arrange(N,prop) %>%
mutate(i = 1:n())
) %>%
ggplot(aes(prop, prop_soft)) +
geom_point() +
facet_wrap(~N) +
geom_abline()

pairs softmax - no jacobian

pairs source model

Then, when I add the simple jacobian everything goes crazy.