Hi everyone,
I’m new to bayesian inference, especially running MCMC for parameter inference. So, I have successfully run my model, but I do not think that I got the result as I expected (see the result below, I have 11 theta, and 1 sigma parameters, all of them do not have good n_eff and R_hat values).
mean se_mean sd 2.50% 25% 50% 75% 97.50% n_eff Rhat
theta[1] 1.99 0.97 0.97 1.02 1.02 1.99 2.96 2.96 1 1.6e4
theta[2] 0.9 0.27 0.27 0.63 0.63 0.9 1.17 1.18 1 9874.4
theta[3] 10.0 8.9e-7 2.3e-5 10.0 10.0 10.0 10.0 10.0 675 1.0
theta[4] 3.08 1.63 1.63 1.44 1.44 3.08 4.71 4.71 1 1.4e4
theta[5] 2.22 1.45 1.45 0.76 0.76 2.22 3.67 3.67 1 1.7e4
theta[6] 0.14 4.6e-4 4.6e-4 0.14 0.14 0.14 0.14 0.14 1 118.71
theta[7] 0.52 0.19 0.19 0.33 0.33 0.52 0.71 0.71 1 7263.1
theta[8] 0.74 0.26 0.26 0.48 0.48 0.74 1.0 1.0 1 6272.7
theta[9] 3.0e-5 3.1e-5 1.4e-4 1.8e-6 2.8e-6 4.3e-6 8.3e-6 2.8e-4 20 1.07
theta[10] 1.09 0.9 0.9 0.19 0.19 1.09 1.99 1.99 1 2.3e4
theta[11] 0.43 0.24 0.24 0.19 0.19 0.43 0.67 0.67 1 1.7e4
sigma 288.66 0.27 0.61 287.67 288.14 288.64 289.22 289.65 5 2.23
lp__ -8.0e6 2.0e4 2.7e4 -8.0e6 -8.0e6 -8.0e6 -7.9e6 -7.9e6 2 1.59
Now, it seems the problem came from incorrect parameter initialization, but honestly I have no idea how I can determine these (I was just using uniform distribution with arbitrary ranges, as follows):
theta[1] ~ uniform(0, 1e1);
theta[2] ~ uniform(0, 1e1);
theta[3] ~ uniform(0, 1e1);
theta[4] ~ uniform(0, 1e2);
theta[5] ~ uniform(0, 1e2);
theta[6] ~ uniform(0, 1e-4);
theta[7] ~ uniform(0, 1e-1);
theta[8] ~ uniform(0, 1e0);
theta[9] ~ uniform(0, 1e5);
theta[10] ~ uniform(0, 1e2);
theta[11] ~ uniform(0, 4);
sigma ~ normal(0, 0.1);
I have tried tweaking the sampling parameters, such as iter, warmup, thin, and adapt_delta but none of these seem to work. I am just confused what kind of information that I can get out of this result that can guide me to improve the model. Can anyone tell me, typically, when improving a MCMC model what do guys look into? Because right now, I am just tweaking every possible number without any idea how that will hypothetically improve my model, and then I ended up spending multiple hours of try and error without any promising result. If anyone can share your rule of thumb in doing model parameter estimation with MCMC, that will be great too.
I also tried looking into parameters correlation and the trace, and I am also confused how to interpret them. If any of these supposed to help, I have attached them below as well (only part of the trace plot, because they seem to have similar trend like these, there were 2 chains, upper part for the first chain and lower part for the second one). The only thing that I noticed is that the behavior of these 2 chains are significantly different, but I do not know why.
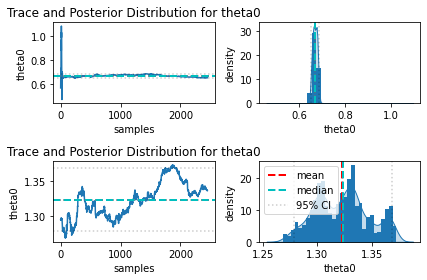
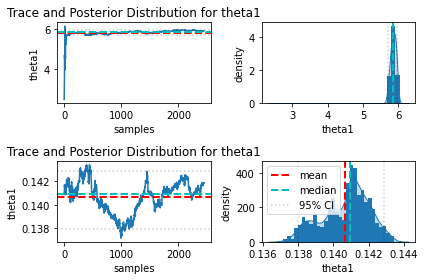


Finally, if it helps, below is my model code:
functions {
real hill_activation(real x, real K, real n) {
real hill;
hill = pow(x, n) / (pow(K, n) + pow(x, n));
return hill;
}
real growth_rate(real y, real a, real b) {
real growth;
growth = (a * (1 - (y/b)));
return growth;
}
real[] gate_model(real t, real[] y, real[] theta, real[] x_r, int[] x_i) {
real OD = y[1];
real ECFn = y[2];
real ECFc = y[3];
real ECF = y[4];
real GFP = y[5];
real bn = theta[1];
real bc = theta[2];
real bg = theta[3];
real syn_ECFn = theta[4];
real syn_ECFc = theta[5];
real syn_ECF = theta[6];
real deg = theta[7];
real syn_GFP = theta[8];
real deg_GFP = theta[9];
real K = theta[10];
real n = theta[11];
real alpha = x_r[1];
real beta = x_r[2];
int ind1 = x_i[1];
int ind2 = x_i[2];
real gamma = growth_rate(OD, alpha, beta);
real dOD = gamma * OD;
real dECFn = bn + syn_ECFn * ind1 - (deg + gamma) * ECFn;
real dECFc = bc + syn_ECFc * ind2 - (deg + gamma) * ECFc;
real dECF = syn_ECF * ECFn * ECFc - (deg + gamma) * ECF;
real dGFP = bg + syn_GFP * hill_activation(ECF, K, n) - (deg_GFP + gamma) * GFP;
return { dOD, dECFn, dECFc, dECF, dGFP };
}
}
data {
int<lower=1> T;
int<lower=1> num_states;
real y0[num_states];
real y[T, 1];
real t0;
real ts[T];
real od_params[2];
int inducers[2];
}
transformed data {
real x_r[2];
int x_i[2];
x_r[1] = od_params[1];
x_r[2] = od_params[2];
x_i[1] = inducers[1];
x_i[2] = inducers[2];
}
parameters {
real<lower=0> theta[11];
real<lower=0> sigma;
}
model {
real y_hat[T, num_states];
theta[1] ~ uniform(0, 1e1);
theta[2] ~ uniform(0, 1e1);
theta[3] ~ uniform(0, 1e1);
theta[4] ~ uniform(0, 1e2);
theta[5] ~ uniform(0, 1e2);
theta[6] ~ uniform(0, 1e-4);
theta[7] ~ uniform(0, 1e-1);
theta[8] ~ uniform(0, 1e0);
theta[9] ~ uniform(0, 1e5);
theta[10] ~ uniform(0, 1e2);
theta[11] ~ uniform(0, 4);
sigma ~ normal(0, 0.1);
y_hat = integrate_ode_rk45(gate_model, y0, t0, ts, theta, x_r, x_i);
y[,1] ~ normal(y_hat[,5], sigma);
}
Thanks a lot.

