as_draws_df() might be helpful here, as it generates a data frame of parameter draws from the model. You can can then manipulate it in R to generate the particular effect you’re interested in. I’m attaching an R script to illustrate this idea. My code gets VERY hairy very quickly, so I apologize if anything is unclear. Hopefully it’s helpful.
ordered_effects.R (8.9 KB)
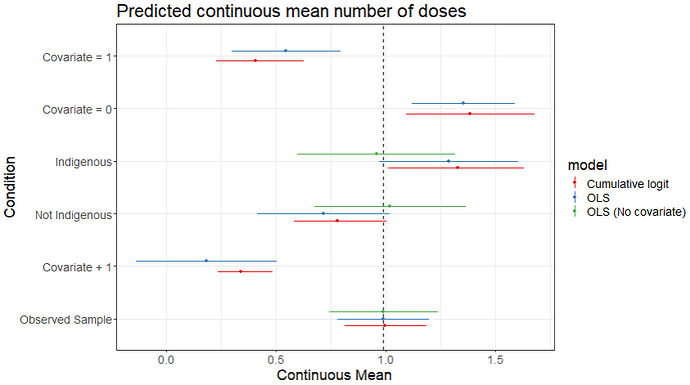
I generate a sample with a binary indicator for indigenous status, a correlated continuous covariate, and an ordered number of doses. I set the parameters so that indigenous status and the covariate both have large effects but they are correlated such that the observed difference between indigenous and non-indigenous samples is negligible.
I estimate the model, then use those draws to estimate different quantities of interest. For example, we could calculate the mean number of doses for the unmodified/observed sample. Or, we could see how that mean changes if we add 1 to the covariate. etc.. We can compare these predicted means to the OLS estimates as well, both including and excluding the covariate in the model.
- Observed Sample: mean number of doses using the unmodified/observed sample
- Covariate + 1: # of doses after adding 1 to the covariate for all cases in the sample
- Not indigenous: Assume everyone in the same is not indigenous while keeping the covariate at the observed level
- Indigenous: same thing but assume everyone is indigenous
- Covariate = 0: Assume everyone has a value of 0 on the covariate while keeping their indigenous status at the observed level
- Covariate = 1: same thing but set the covariate to 1 for all cases
Finally, we can calculate the conditional effect sizes by comparing these means. For example, the effect of adding 1 to the covariate would be the difference between the adjusted mean and the predicted mean for the observed sample.