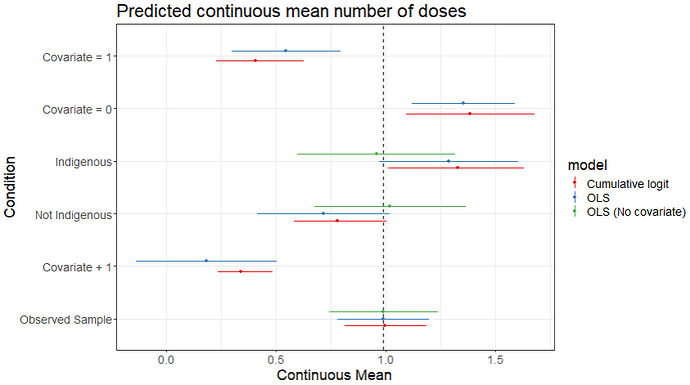
Hi there,
I am modelling the effects of age on the number of treatment doses over 3 months. Due to the shape of this distribution, I’m finding that the model is best fit using ordinal regression (see an example of the code below). A problem however, is that I wish to report the results on a continuous scale, like one would with linear regression (e.g. 1 year increase in age is associated with a mean increase of 2.5 doses).
brm(ndose ~ age, family=cumulative(),iter=10000,chains=4,cores=4,data=ndose_met)
I cannot find a way to back transform the results to a continuous scale. The conditional_effects() function from brms (with categorical=FALSE) seems to be giving me what I want, but the output is a plot rather than the mean difference + credible interval.
Does anyone know how to do this (either within R or Stan)? I have managed to calculate the mean difference in Stan if the predictor is categorical (e.g. sex), but not continuous (e.g. age).
The code that I have used to calculate the mean difference for the categorical variable indigenous_status (I’ve taken the brms base code and added in more to the generated quantities section. Apologies for the messy workarounds I’ve done.)
// generated with brms 2.21.0
functions {
/* cumulative-logit log-PDF for a single response
* Args:
* y: response category
* mu: latent mean parameter
* disc: discrimination parameter
* thres: ordinal thresholds
* Returns:
* a scalar to be added to the log posterior
*/
real cumulative_logit_lpmf(int y, real mu, real disc, vector thres) {
int nthres = num_elements(thres);
if (y == 1) {
return log_inv_logit(disc * (thres[1] - mu));
} else if (y == nthres + 1) {
return log1m_inv_logit(disc * (thres[nthres] - mu));
} else {
return log_inv_logit_diff(disc * (thres[y] - mu), disc * (thres[y - 1] - mu));
}
}
/* cumulative-logit log-PDF for a single response and merged thresholds
* Args:
* y: response category
* mu: latent mean parameter
* disc: discrimination parameter
* thres: vector of merged ordinal thresholds
* j: start and end index for the applid threshold within 'thres'
* Returns:
* a scalar to be added to the log posterior
*/
real cumulative_logit_merged_lpmf(int y, real mu, real disc, vector thres, array[] int j) {
return cumulative_logit_lpmf(y | mu, disc, thres[j[1]:j[2]]);
}
/* ordered-logistic log-PDF for a single response and merged thresholds
* Args:
* y: response category
* mu: latent mean parameter
* thres: vector of merged ordinal thresholds
* j: start and end index for the applid threshold within 'thres'
* Returns:
* a scalar to be added to the log posterior
*/
real ordered_logistic_merged_lpmf(int y, real mu, vector thres, array[] int j) {
return ordered_logistic_lpmf(y | mu, thres[j[1]:j[2]]);
}
}
data {
int<lower=1> N; // total number of observations
array[N] int Y; // response variable
int<lower=2> nthres; // number of thresholds
int<lower=1> K; // number of population-level effects
matrix[N, K] X; // population-level design matrix
int<lower=1> Kc; // number of population-level effects after centering
int prior_only; // should the likelihood be ignored?
}
transformed data {
matrix[N, Kc] Xc; // centered version of X
vector[Kc] means_X; // column means of X before centering
for (i in 1:K) {
means_X[i] = mean(X[, i]);
Xc[, i] = X[, i] - means_X[i];
}
int n_indigenous = 0;
for(i in 1:N){
if(X[i,1] == 1) n_indigenous += 1;
}
int n_notindigenous = N - n_indigenous;
}
parameters {
vector[Kc] b; // regression coefficients
ordered[nthres] Intercept; // temporary thresholds for centered predictors
}
transformed parameters {
real disc = 1; // discrimination parameters
real lprior = 0; // prior contributions to the log posterior
lprior += student_t_lpdf(Intercept | 3, 0, 2.5);
}
model {
// likelihood including constants
if (!prior_only) {
target += ordered_logistic_glm_lpmf(Y | Xc, b, Intercept);
}
// priors including constants
target += lprior;
}
generated quantities {
// compute actual thresholds
vector[nthres] b_Intercept = Intercept + dot_product(means_X, b);
array[N] int yhat;
real mu_ind = 0;
real mu_notind = 0;
real n_indigenous2 = n_indigenous;
real n_notindigenous2 = n_notindigenous;
for(i in 1:N){
yhat[i] = ordered_logistic_rng( Xc[i,] * b, Intercept);
if(X[i,1] == 1) mu_ind += yhat[i]/n_indigenous2;
else mu_notind += yhat[i]/n_notindigenous2;
}
real RR = mu_ind/mu_notind;
real MD = mu_ind - mu_notind;
}