Hello,
Sorry in advance for the long post…
I am working to fit an ordinal logistic regression with brms to species’ risk extinction categories (ranging in order from Low to High risk : 5 levels) vs several predictors (numeric, nominal, ordered; ecological characteristics). I have around 100 observations, but some levels are not observed in all extinction categories.
I have read quite a lot of posts here and some other docs but I would like some guidance as I have trouble making sense of the prior predictive checks and model checking. I have no real a priori of what the Intercept thresholds or predictor relations should be.
My final aim is to use the package projpred to do some predictor selection as I expect that some factors may be redundant.
Starting with the NULL model below:
brm(
formula = extinction_cat ~ 1
, data = df_extinction
, family = cumulative("logit")
# , sample_prior = 'only'
, iter = 18000
, warmup = 12000
, thin = 3
, chains = 3
, cores = 3
)
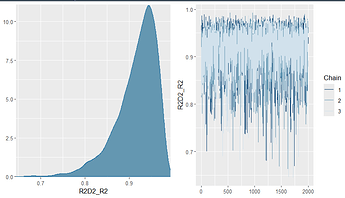
I get prior-data conflict on one of the Intercept with priorsense::powerscale_sensitivity

I get these posterior pp_check
And this when I run it with sample_prior = 'only':
I found that using slightly weaker prior prior = c(prior(student_t(3, 0, 5), class = "Intercept")) remove the prior-data conflict but seems to increase the dome-shaped of the pp_check.
(1) Is this the right decision?
Then I add one nominal predictor to the model. This predictor is not really well distributed in each extinction categories and two levels have low observation count and only in one extinction category (only 2 species in level C, and 4 in F).
When checking the posteriors, one dietC has a large distribution and powerscale_sensitivity indicates a weak likelihood (which I guess makes sense as there is not a lot of info in the observations).
bars_grouped pp_check:
(2) Why is dietC completely off? And why dietF isn’t then?
As prior_sample = “only” doesn’t work with the default priors, I set the prior of b to the same weakly informative prior as for the Intercept student_t(3, 0, 5) which reduce the uncertainty in dietC but the weak likelihood is still there (and pp_check looks the same).

with sample_prior = 'only':
With power scaling sensitivity (powerscale_plot_ecdf) :
If I precise the priors on b student_t(3, 0, 2.5), it goes to prior-data conflict

(3) Not sure what the solution is here for the final model between choosing prior-data conflict or weak likelihood.
Thanks for your help!
- Operating System: Windows
- brms Version: 2.20.4
- priorsense Version: 0.0.0.9