- Operating System: iOS 10.13.5
- brms Version: 2.6.0
TL;DR
I have a binomial response variable that is a function of five continuous covariates, but I need to account for temporal autocorrelation with an auto-regressive structure. The only way to do that is by first taking the binomial response variable, set it as a number and logit-transform it, fit the model with a Gaussian distribution, and then add the auto-regressive structure (cor_ar). This paper did the same thing with the lme{nlme} function, but there are mixed opinions about doing that kind of transformation. Such a thing can be done with brms (see below for reproducible example), but I was wondering whether it’s the right thing to do and what the opinions are with doing such a transformation (again, see below, along with other queries about weights).
The data set I’m working on is only slightly different from this reproducible example. Let’s say we have a dataset with a binomial response variable, a continuous covariate, and a sequential time covariate.
library(brms)
library(lubridate)
library(car)
library(bayesplot)
####Simulate data
#Set the seed
RNGkind('Mersenne-Twister')
set.seed(42)
#Create covariate x
x <- rnorm(1000)
#Create coefficients
beta0 <- 0.6
beta1 <- 0.7
#Create responses
z <- beta0 + beta1*x
pr <- 1/(1+exp(-z))
y <- rbinom(1000, 1, pr)
#Create timestamps
time <- seq(dmy_hms("01-01-2019 00:00:00"), dmy_hms("01-03-2019 00:00:00"), length.out = 1000)
#Create the data frame
data <- data.frame(y, x, time)
data$y <- factor(data$y, levels = c('0', '1'))
In such a case, you would fit a bernoulli or binomial distribution in a brm model:
#Control model
ctrl.mod <- brm(y ~ x, family = bernoulli(link = 'logit'), data = data,
seed = 42, iter = 10000, warmup = 1000, chain = 4, cores = 4)
But given the different frequency of the binomial response variable…
table(y)
y
0 1
393 607
…we might want to add weights to our model.
#Weighted model
data$weights <- ifelse(y == 1, 1, 1/(table(y)[1]/table(y)[2]))
wt.mod <- brm(y | weights(weights) ~ x, family = bernoulli(link = 'logit'), data = data,
seed = 42, iter = 10000, warmup = 1000, chain = 4, cores = 4)
But we have a time covariate, and we may want to account for temporal autocorrelation with an auto-regressive structure using the cor_ar function. The trouble is that it is only available for the Gaussian distribution.
This paper also sought to do the same thing. To quote from the article:
We logit transformed daily residence probabilities (adding 0.001 to 0 values and subtracting 0.001 from values of 1) and used linear mixed-effects models in the R package ‘nlme’ (Pinheiro et al. 2015), including shark ID as a random effect, and environmental covariates as fixed effects.
So I’ll try and do the same here.
#Unweighted first order AR model
data$logit.res <- logit(ifelse(y == 1, y - 0.01, y + 0.01))
ar.unwt.mod <- brm(logit.res ~ x, family = gaussian(link = 'identity'), data = data,
seed = 42, iter = 10000, warmup = 1000, chain = 4, cores = 4,
autocor = cor_ar(~ time, p = 1, cov = F))
But we’ll create another model with an auto-regressive structure with weights for good measure:
#Weighted first order AR model
ar.wt.mod <- brm(logit.res | weights(weights) ~ x, family = gaussian(link = 'identity'), data = data,
seed = 42, iter = 10000, warmup = 1000, chain = 4, cores = 4,
autocor = cor_ar(~ time, p = 1, cov = F))
The coefficients aren’t so important. What I’m most concerned about are the posterior predictive distributions:
The models on the left are unweighted models, those on the right are weighted. Top row is fitted with a Binomial distribution, bottom row is fitted with a Gaussian distribution with a logit-transformed response variable.
The ctrl.mod has a good fit, but when we include weights, the wt.mod doesn’t fit very well because of the weights. What’s really concerning are the bottom two graphs. In order to fit an auto-regressive structure to account for temporal autocorrelation, I had to logit-transform the response variable and fit a Guassian distribution, which is reflected in the posterior predictive distribution. But because I fitted the auto-regressive structure, the fit really suffers; the mean of the posterior distribution is hanging in between the two observed values found in the data, differing due to the weights (or lack-thereof) of the response variable.
The LOO-PIT plots are pretty wild as well:
Same as previous figure
What’s happening in the top two graphs is beyond me. The bottom graphs look typical of a non-normal distribution, which make sense since the data isn’t normally distributed anyway.
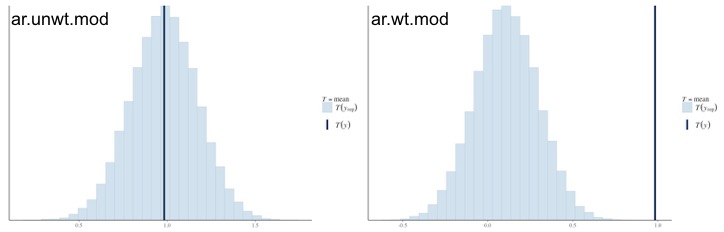
Mean recoveries are also varied (I could only get mean recoveries for the AR models, the two binomial models kept returning polygon edge not found as an error):
The unweighted AR model had decent mean recovery, but the weighted AR model was way off.
This answer to this question said that logit-transforming the response variable is not advised, though a comment to the answer said that this type of transformation is very common (such as the previously linked paper). Hence why I thought it’d be all right to do this with brms, especially since I need to account for temporal autocorrelation. But then again, I’m not sure.
Although this is a simulated dataset, the bayesplot presented here exactly the same plots I’m getting for my dataset.
My questions are:
- Is it more important to ensure the posterior predictive distributions fit the observed distribution of the data?
- Should weights be used at all if it means the distributions will not align or if mean recoveries are way off? Are imbalances in the response variable generally handled differently within a Bayesian framework? Because it seems adding weights is getting some poor fits all around, so I’m wondering if I should do away with them entirely.
- When we need to account for temporal autocorrelation, is logit-transforming the binomial response variable and specifying a Gaussian distribution the best way to go just so that we can fit an autoregressive structure, even if the distributions don’t align? If not, what’s the best way to account for temporal autocorrelation with a binomial response variable?