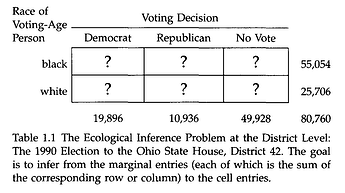
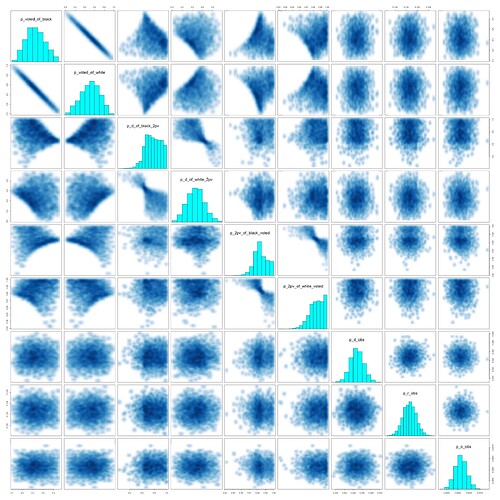
I have been trying to model a basic ecological inference problem in RStan.
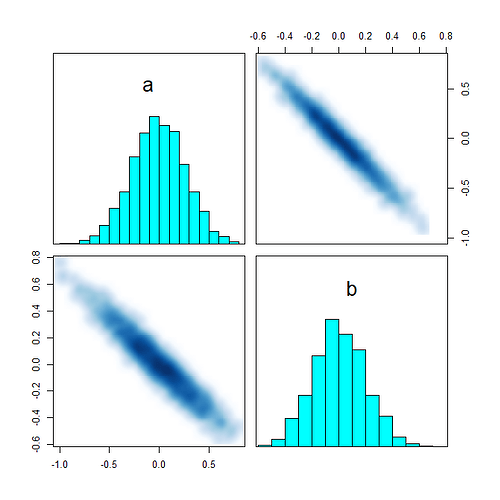
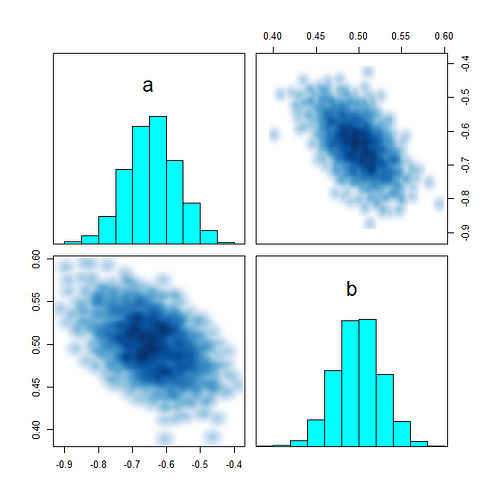
I have introduced a fourth column: third-party votes. I will make a hierarchical model that incorporates data from the precincts that partition the district, but I want to make sure I am doing everything right first. I believe this is a fairly nice parametrization, but I am new to Stan and I don’t really understand how to read the pairs() plots.
This is rstan version 2.32.7 (Stan version 2.32.2).
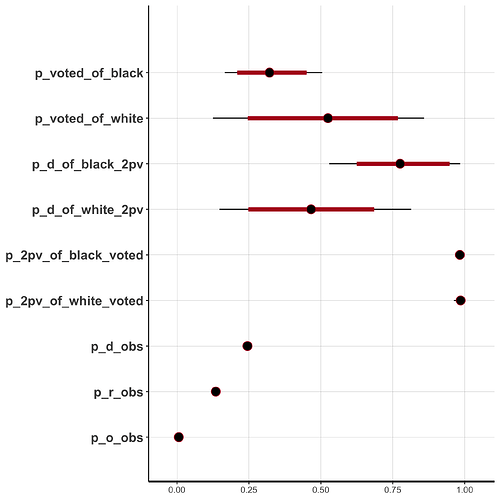
Results
mean se_mean sd 2.5% 25% 50% 75% 97.5% n_eff Rhat
p_voted_of_black 0.33 0.00 0.09 0.17 0.26 0.32 0.39 0.51 1168 1
p_voted_of_white 0.51 0.01 0.19 0.13 0.37 0.52 0.65 0.86 1165 1
p_d_of_black_2pv 0.78 0.00 0.13 0.53 0.68 0.78 0.88 0.98 1314 1
p_d_of_white_2pv 0.47 0.00 0.17 0.15 0.35 0.47 0.58 0.81 1538 1
p_2pv_of_black_voted 0.98 0.00 0.01 0.97 0.98 0.98 0.99 1.00 1650 1
p_2pv_of_white_voted 0.98 0.00 0.01 0.96 0.98 0.99 0.99 1.00 1626 1
p_d_obs 0.24 0.00 0.00 0.24 0.24 0.24 0.25 0.25 4382 1
p_r_obs 0.13 0.00 0.00 0.13 0.13 0.13 0.14 0.14 3762 1
p_o_obs 0.01 0.00 0.00 0.01 0.01 0.01 0.01 0.01 3866 1
(no warnings)
Stan code
data {
int<lower=1> n; // total number of people in the district
int<lower=0, upper=n>
d, r, o, // Democratic votes, Republican votes, third-party votes (non-voters inferred)
black; // (white population inferred)
// parameters for the (beta distribution) priors of
// P(voted|black), P(voted|white),
// P(D|black,voted D or R), P(D|white,voted D or R),
// P(D or R|black,voted), P(D or R|white, voted)
array[6] real<lower=0> alphas;
array[6] real<lower=0> betas;
}
transformed data {
real<lower=0, upper=1>
p_black = (black + 0.0) / n,
p_white = 1 - p_black;
}
parameters {
real<lower=0, upper=1>
p_voted_of_black, // P(voted|black)
p_voted_of_white, // P(voted|white)
p_d_of_black_2pv, // P(D|voted D or R,black)
p_d_of_white_2pv, // P(D|voted D or R,white)
p_2pv_of_black_voted, // P(D or R|black,voted)
p_2pv_of_white_voted; // P(D or R|white,voted)
}
transformed parameters {
real<lower=0, upper=1>
p_d_obs, // P(D) (estimated from parameters)
p_r_obs, // P(R) (estimated from parameters)
p_o_obs; // P(3rd party) (estimated from parameters)
p_d_obs = p_d_of_black_2pv * p_2pv_of_black_voted * p_voted_of_black * p_black
+ p_d_of_white_2pv * p_2pv_of_white_voted * p_voted_of_white * p_white;
p_r_obs = (1 - p_d_of_black_2pv) * p_2pv_of_black_voted * p_voted_of_black * p_black
+ (1 - p_d_of_white_2pv) * p_2pv_of_white_voted * p_voted_of_white * p_white;
p_o_obs = (1 - p_2pv_of_black_voted) * p_voted_of_black * p_black
+ (1 - p_2pv_of_white_voted) * p_voted_of_white * p_white;
}
model {
// Priors
p_voted_of_black ~ beta(alphas[1], betas[1]);
p_voted_of_white ~ beta(alphas[2], betas[2]);
p_d_of_black_2pv ~ beta(alphas[3], betas[3]);
p_d_of_white_2pv ~ beta(alphas[4], betas[4]);
p_2pv_of_black_voted ~ beta(alphas[5], betas[6]);
p_2pv_of_white_voted ~ beta(alphas[6], betas[6]);
d ~ binomial(n, p_d_obs);
r ~ binomial(n, p_r_obs);
o ~ binomial(n, p_o_obs);
}
R code
# pp. 13 of King
library(rstan)
library(ggplot2)
seed <- 1
set.seed(seed)
o <- 500 # 3rd party votes
n <- 80760 + o
d <- 19896
r <- 10936
nv <- 49928
black <- 55054 + o
white <- 25706
stopifnot(d + r + o + nv == n, black + white == n)
prior_alphas <- c(2, 2, 4, 2.5, 100, 100)
prior_betas <- c(2, 2, 1, 2.5, 1, 1)
data <- list(n = n, d = d, r = r, o = o, black = black, alphas = prior_alphas, betas = prior_betas)
fit <- stan(
file = "ohio-state-house-42.stan",
data = data,
chains = 4,
warmup = 1000,
iter = 2000,
cores = 4,
seed = seed
)
pars <- c("p_voted_of_black", "p_voted_of_white",
"p_d_of_black_2pv", "p_d_of_white_2pv",
"p_2pv_of_black_voted", "p_2pv_of_white_voted",
"p_d_obs", "p_r_obs", "p_o_obs")
print(fit, pars = pars)
plot(fit, pars = pars)
ggsave("ohio-state-house-42.png")
png("ohio-state-house-42-pairs.png", height = 2000, width = 2000)
suppressWarnings(pairs(fit, pars = pars))
dev.off()