@avehtari, thank you so much. I have changed the model as you suggested and that fixed the problem completely! Model comparison now strongly prefers the two-gaussian model, as it should (I did many other tests with synthetic data as well). WAIC and PSIS numbers are almost identical. Pareto K values are all below 0.5.
The new model also samples much faster, with no Stan warnings and higher effective number of samples, so the new model is a better solution to my divergent transitions question.
I’ve also checked that the fixed model produces distributions of model parameters that are almost identical to those from the previous model. So the old and new models are probably mathematically equivalent.
Here is the fixed versions of the models and the plots.
Model 1: single gaussian
// Single Gaussian population model
data {
int<lower=1> n; // Number of data points
real y[n]; // Observed values
real uncertainties[n]; // Uncertainties
real sigma_stdev_prior;// Prior for standard deviation of population spread
}
parameters {
real mu; // Population location
real<lower=0> sigma; // Population spread
}
model {
// Set priors
sigma ~ normal(0, sigma_stdev_prior);
mu ~ normal(0, 1);
// Loop through observed values and calculate their probabilities
// for a Gaussian distribution, accounting for uncertainty of each measurment
for (i in 1:n) {
target += normal_lpdf(y[i] | mu, sqrt(sigma^2 + uncertainties[i]^2));
}
}
generated quantities{
vector[n] lpd_pointwise;
for (i in 1:n) {
lpd_pointwise[i] = normal_lpdf(y[i] | mu, sqrt(sigma^2 + uncertainties[i]^2));
}
}
Model 1: mixture of two gaussians
// Guassian mixture model
data {
int<lower=1> n; // Number of data points
real y[n]; // Observed values
real uncertainties[n]; // Uncertainties
real sigma_stdev_prior;// Prior for standard deviation of population spread
}
parameters {
real<lower=0,upper=1> r; // Mixing proportion
ordered[2] mu; // Locations of mixture components
real<lower=0> sigma; // Spread of mixture components
}
model {
// Set priors
sigma ~ normal(0, sigma_stdev_prior);
mu[1] ~ normal(0, 1);
mu[2] ~ normal(0, 1);
r ~ beta(2, 2);
// Loop through observed values
// and mix two Gaussian components accounting for uncertainty of each measurment
for (i in 1:n) {
target += log_mix(1 - r,
normal_lpdf(y[i] | mu[1], sqrt(sigma^2 + uncertainties[i]^2)),
normal_lpdf(y[i] | mu[2], sqrt(sigma^2 + uncertainties[i]^2)));
}
}
generated quantities{
vector[n] lpd_pointwise;
for (i in 1:n) {
lpd_pointwise[i] = log_mix(1 - r,
normal_lpdf(y[i] | mu[1], sqrt(sigma^2 + uncertainties[i]^2)),
normal_lpdf(y[i] | mu[2], sqrt(sigma^2 + uncertainties[i]^2)));
}
}
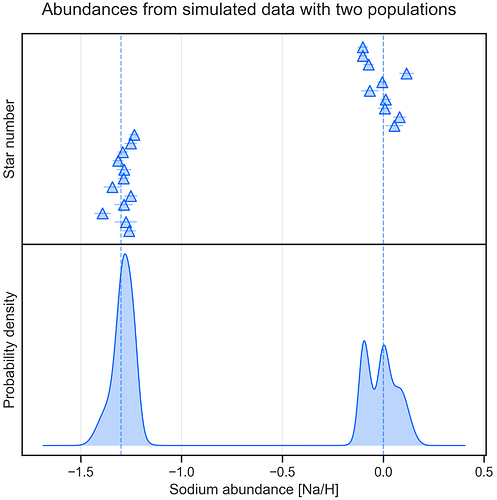
Figure 10: Synthesised observations
Table 1: WAIC output
| WAIC | SE | dWAIC | dSE | pWAIC | Weight | |
|---|---|---|---|---|---|---|
| Two populations | -5.06 | 6.81 | 3.67 | 1.00 | ||
| One population | 91.18 | 4.28 | 96.24 | 5.37 | 3.82 | 0.00 |
Table 2: PSIS output
| PSIS | SE | dPSIS | dSE | pPSIS | MaxK | Weight | |
|---|---|---|---|---|---|---|---|
| Two populations | -4.88 | 6.89 | 3.76 | 0.49 | 1.00 | ||
| One population | 91.20 | 4.28 | 96.08 | 5.43 | 3.83 | 0.19 | 0.00 |
Figure 11: Comparing models with WAIC
Figure 12: Comparing models with PSIS
Figure 13: Pareto K values
Full Code
The full code to compare the models and make the above plots is located here: