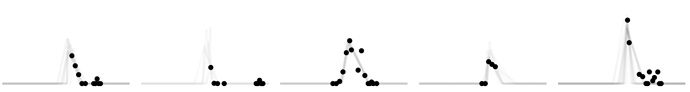
Hi everyone - I’m running into some issues with implementing a non-centered version of a model I’ve implemented. I have data that’s described by a piecewise linear model: it’s flat, then increases to a peak, then decreases back to the original level and flattens again.
I’ve implemented a hierarchical model that fits the peak time (‘tp’), the gap between the first intercept and the peak (‘wp’), the gap between the peak and the second intercept (‘wr’), and the size of the peak (‘dp’) for each of 40 individuals based on a population mean and standard deviation for each parameter.
The centered version works (a few traces below), but throws some divergences which I’d like to clear up.
I tried to implement a non-centered version. It gives equally good fits to the data - but many many more divergences and the chains don’t mix well.
Any ideas what might be going on here? Thanks so much in advance! Relevant code snippets pasted below.
Centered version:
parameters {
real<lower=0, upper=lod> dpmean;
real<lower=0, upper=wpmax> wpmean;
real<lower=0, upper=wrmax> wrmean;
real<lower=0> dpsd;
real<lower=0> wpsd;
real<lower=0> wrsd;
real tp[n_id];
real<lower=0, upper=lod> dp[n_id];
real<lower=0, upper=wpmax> wp[n_id];
real<lower=0, upper=wrmax> wr[n_id];
// real dpraw[n_id];
// real wpraw[n_id];
// real wrraw[n_id];
}
transformed parameters {
real process_sd[N];
real mu[N];
// real<lower=0, upper=lod> dp[n_id];
// real<lower=0, upper=wpmax> wp[n_id];
// real<lower=0, upper=wrmax> wr[n_id];
// Non-centering:
// for(i in 1:n_id){
// dp[i] = dpmean + dpsd*dpraw[i];
// wp[i] = wpmean + wpsd*wpraw[i];
// wr[i] = wrmean + wrsd*wrraw[i];
// }
for(i in 1:N){
mu[i]=mufun(t[i], tp[id[i]], wp[id[i]], wr[id[i]], dp[id[i]]);
process_sd[i]=sqrt((sigma*sigma) + (epsilon[i]*epsilon[i]));
};
}
model {
// Hyperparameter priors
dpmean ~ normal(dpmean_prior,dpsd_prior) T[0,lod];
dpsd ~ cauchy(0,1) T[0,];
wpmean ~ normal(wpmean_prior, wpsd_prior) T[0,wpmax];
wpsd ~ cauchy(0,1) T[0,];
wrmean ~ normal(wrmean_prior, wrsd_prior) T[0,wrmax];
wrsd ~ cauchy(0,1) T[0,];
// Individual-level sampling statements:
tp ~ normal(0,tpsd);
// Centered: -----
for(i in 1:n_id){
dp[i] ~ normal(dpmean, dpsd) T[0, lod];
wp[i] ~ normal(wpmean, wpsd) T[0, wpmax];
wr[i] ~ normal(wrmean, wrsd) T[0, wrmax];
}
// Non-centered: -----
// for(i in 1:n_id){
// dpraw[i] ~ normal(0,1) T[-dpmean/dpsd, (lod-dpmean)/dpsd];
// wpraw[i] ~ normal(0,1) T[-wpmean/wpsd, (wpmax-wpmean)/wpsd];
// wrraw[i] ~ normal(0,1) T[-wrmean/wrsd, (wrmax-wrmean)/wrsd];
// }
// Main model specification:
for(i in 1:N){
target += log_sum_exp(
log1mlambda + normal_lpdf(ydrop[i] | mu[i], process_sd[i]),
loglambda + exponential_lpdf(ydrop[i] | 1/fpmean));
if (ydrop[i] < 0 || ydrop[i] > lod)
target += negative_infinity();
else
target += -log_diff_exp(
log1mlambda + normal_lcdf(lod | mu[i], process_sd[i]),
log1mlambda + normal_lcdf(0 | mu[i], process_sd[i]));
// see https://mc-stan.org/docs/2_18/reference-manual/sampling-statements-section.html
}
}
Non-centered version:
parameters {
real<lower=0, upper=lod> dpmean;
real<lower=0, upper=wpmax> wpmean;
real<lower=0, upper=wrmax> wrmean;
real<lower=0> dpsd;
real<lower=0> wpsd;
real<lower=0> wrsd;
real tp[n_id];
// real<lower=0, upper=lod> dp[n_id];
// real<lower=0, upper=wpmax> wp[n_id];
// real<lower=0, upper=wrmax> wr[n_id];
real dpraw[n_id];
real wpraw[n_id];
real wrraw[n_id];
}
transformed parameters {
real process_sd[N];
real mu[N];
real<lower=0, upper=lod> dp[n_id];
real<lower=0, upper=wpmax> wp[n_id];
real<lower=0, upper=wrmax> wr[n_id];
// Non-centering:
for(i in 1:n_id){
dp[i] = dpmean + dpsd*dpraw[i];
wp[i] = wpmean + wpsd*wpraw[i];
wr[i] = wrmean + wrsd*wrraw[i];
}
for(i in 1:N){
mu[i]=mufun(t[i], tp[id[i]], wp[id[i]], wr[id[i]], dp[id[i]]);
process_sd[i]=sqrt((sigma*sigma) + (epsilon[i]*epsilon[i]));
};
}
model {
// Hyperparameter priors
dpmean ~ normal(dpmean_prior,dpsd_prior) T[0,lod];
dpsd ~ cauchy(0,1) T[0,];
wpmean ~ normal(wpmean_prior, wpsd_prior) T[0,wpmax];
wpsd ~ cauchy(0,1) T[0,];
wrmean ~ normal(wrmean_prior, wrsd_prior) T[0,wrmax];
wrsd ~ cauchy(0,1) T[0,];
// Individual-level sampling statements:
tp ~ normal(0,tpsd);
// Centered: -----
// for(i in 1:n_id){
// dp[i] ~ normal(dpmean, dpsd) T[0, lod];
// wp[i] ~ normal(wpmean, wpsd) T[0, wpmax];
// wr[i] ~ normal(wrmean, wrsd) T[0, wrmax];
// }
// Non-centered: -----
for(i in 1:n_id){
dpraw[i] ~ normal(0,1) T[-dpmean/dpsd, (lod-dpmean)/dpsd];
wpraw[i] ~ normal(0,1) T[-wpmean/wpsd, (wpmax-wpmean)/wpsd];
wrraw[i] ~ normal(0,1) T[-wrmean/wrsd, (wrmax-wrmean)/wrsd];
}
// Main model specification:
for(i in 1:N){
target += log_sum_exp(
log1mlambda + normal_lpdf(ydrop[i] | mu[i], process_sd[i]),
loglambda + exponential_lpdf(ydrop[i] | 1/fpmean));
if (ydrop[i] < 0 || ydrop[i] > lod)
target += negative_infinity();
else
target += -log_diff_exp(
log1mlambda + normal_lcdf(lod | mu[i], process_sd[i]),
log1mlambda + normal_lcdf(0 | mu[i], process_sd[i]));
// see https://mc-stan.org/docs/2_18/reference-manual/sampling-statements-section.html
}
}