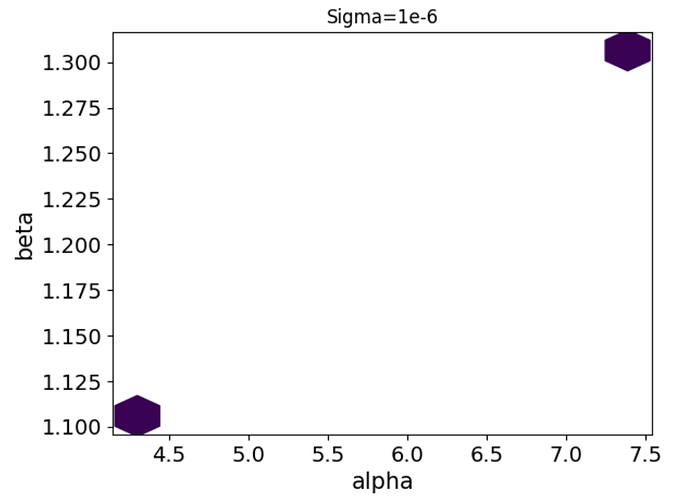
Hi,
I’m an absolute newbie to the forums and fairly new to Stan + Bayesian Modeling, so please forgive the noobness of this question.
In a nutshell: I run into unexpected behavior when adding normally distributed error to an inverse-gamma distributed random variable. I’m almost certain this is because of misspecification (in line with the warnings I’m receiving) but I’d love some pointers as to why this is the case.
I’m simulating inverse-gamma distributed values (using PyStan) with
sim_code = """
data {
int<lower=1> N;
}
generated quantities {
real y[N];
for (i in 1:N)
y[i] = inv_gamma_rng(30, 300);
}
"""
sim = pystan.StanModel(model_code=sim_code).sampling(
data={'N': 100},
chains=1,
iter=1,
seed=17,
warmup=0,
algorithm='Fixed_param'
)
As expected I can recover alpha=30, beta=300 perfectly with
data {
int<lower=1> N;
real y[N];
}
parameters {
real<lower=0> alpha;
real<lower=0> beta;
}
model {
y ~ inv_gamma(alpha, beta);
}
However, if I assume there’s additional normally distributed error, I receive E-BFMI warnings, a handful of divergences, and a weirdly bi-modal distribution of alphas and betas which is way off. Here’s the code I’m using:
data {
int<lower=1> N;
real y[N];
}
transformed data {
real sigma = .001;
}
parameters {
real<lower=0> alpha;
real<lower=0> beta;
real<lower=0> noise_free_y[N];
}
model {
noise_free_y ~ inv_gamma(alpha, beta);
y ~ normal(noise_free_y, sigma);
}
Of course, there is no normally distributed error in my simulation but my understanding would be that wrapping the inverse-gamma distribution with a normal distribution (here with marginal variance) should not corrupt the model. What am I missing?
Few notes:
- I deliberately left out priors here to keep things brief, in this simple example I would expect negligible impact
- In case you’re wondering why I’m considering adding normally distributed error: I’m planning to regress on
yusing additional variables linearly summed with the inverse-gamma random variables - I’m aware of this but feels this goes deeper than needed at the moment
Thanks in advance for engaging on this basic matter!