I am analysing an experiment looking at abstinence rates among participants in a clinical drug and alcohol trial. There were two groups, those who received the new treatment and those who received placebo. Abstinence during the previous week was measured at 0 (baseline), 4, 8, 12, and 24 weeks. There was a high rate of attrition/missing data so that of the 128 that commenced the trial, only 55 supplied data at follow-up. I want to estimate the group difference in odds of abstinence at each time point, controlling for the other time points. I am especially interested in the difference in odds of abstinence at week 24 (controlling for all the other timepoints).
Here is the data.
df <- data.frame(id = factor(rep(1:128,each=5)),
time = factor(rep(c(0,4,8,12,24),times=128)),
group = factor(c(rep("placebo",335),rep("treatment",305))),
abs = c(0, 0, 0, 0, 0, 0, NA, NA, NA, NA, 0, NA, NA, NA, NA, 0, 1, 1, NA, 1, 0, 0, 0, NA, NA, 0, 0, 1, NA, 0, 0, 0, NA, NA, 1, 0, NA, NA, NA, 0, 0, 0, NA, NA, NA, 0, NA, NA, NA, 1, 0, 0, NA, NA, NA, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, NA, NA, NA, 0, NA, NA, NA, NA, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, NA, NA, 0, 0, NA, NA, NA, NA, 0, 0, 0, 0, 0, 0, 0, 0, 0, NA, 0, 0, 0, 0, NA, 0, NA, NA, NA, NA, 0, 0, NA, NA, NA, 0, 0, NA, 0, 0, 0, 0, 0, 0, NA, 0, 0, 0, 0, 0, 0, 0, 0, 0, NA, 0, 0, NA, NA, NA, 0, 0, 0, 0, 0, 0, 0, 0, 0, NA, 0, 0, NA, 0, 0, 0, 0, 0, NA, 0, 0, 0, 0, 0, NA, 0, 0, 0, 0, 0, 0, 0, 0, 0, NA, 0, NA, NA, NA, NA, 0, 0, 0, NA, NA, 0, 1, 1, 1, NA, 0, 0, 0, 0, NA, 0, 0, 0, NA, NA, 0, 0, 0, 0, NA, 0, 0, 0, 0, 0, 0, NA, NA, NA, NA, 0, 0, NA, NA, NA, 0, 0, 1, 1, NA, 0, 1, 1, 1, 0, 0, 0, 0, 0, 0, 0, 1, 1, 1, 0, 0, 0, 0, 0, NA, 0, 0, 1, 1, 1, 0, 0, 0, NA, NA, 0, 0, 0, 0, NA, 0, 0, 0, 0, 1, 0, 0, 1, 1, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, NA, 0, 0, NA, NA, NA, 0, 0, 0, 0, 0, 0, NA, NA, NA, NA, 0, 1, 1, 1, 1, 0, 0, 0, 0, 0, 0, 0, NA, NA, NA, 0, NA, NA, NA, NA, 0, 0, 0, 0, NA, 0, 0, 0, 0, NA, 0, NA, NA, NA, NA, 0, 0, 0, 0, 1, 0, 1, NA, NA, 0, 0, 0, 0, NA, 0, 0, 0, 0, 0, 0, 0, 0, 0, NA, NA, 0, NA, NA, NA, NA, 0, NA, NA, NA, NA, 0, NA, NA, NA, NA, 0, 0, 0, 0, 1, 0, 0, 1, 0, 1, 0, 0, 0, 0, 0, 0, 0, NA, 0, NA, 0, NA, NA, NA, NA, 0, 0, 0, 0, 0, 0, 0, NA, NA, NA, 0, 0, NA, 0, NA, 0, NA, 1, NA, 1, 0, 0, NA, NA, NA, 0, 1, 1, 1, 1, 0, 0, 0, NA, NA, 0, NA, NA, NA, NA, 0, NA, NA, NA, NA, 0, 0, 0, 0, NA, 0, 0, 0, 0, NA, 0, 0, NA, 0, NA, 0, 0, 0, 0, 0, 0, NA, NA, NA, NA, 0, 0, 0, 0, 0, 0, 0, 1, 1, 0, 0, NA, NA, NA, NA, 0, 0, NA, NA, NA, 0, 0, 0, 0, NA, 0, 0, NA, 0, NA, 0, 0, 0, 0, NA, 0, 0, 0, 0, NA, 0, 0, 0, 0, NA, 0, NA, NA, NA, NA, 0, 1, 1, 1, 1, 0, NA, NA, NA, NA, 0, 0, NA, NA, NA, 0, 0, 1, 1, 1, 0, 1, 1, 1, 1, 0, 1, 1, 1, 1, 0, 1, 1, 1, 1, 0, 0, 0, 1, 0, 0, 0, 0, 1, 1, 0, 0, 0, 0, 0, 0, 0, NA, NA, NA, 0, NA, NA, NA, NA, 0, 0, 0, 0, 1, 0, 0, NA, NA, NA, 0, 0, 0, 0, 0, 0, 0, NA, NA, NA, 0, 1, 1, 1, 1, 0, 0, 1, 0, NA, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, NA, 0, NA, 0, 0, NA, 0, NA, 0, 0, 0, 0, NA, 0, 0, NA, NA, NA))
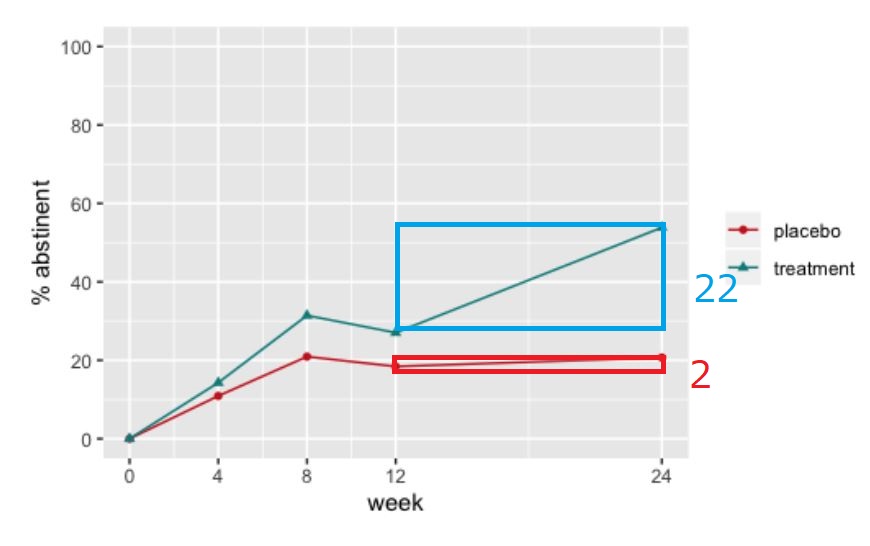
The proportions of the observed cases who were abstinent over time look like this

Which, on face value, looks like a decent treatment effect, especially at week 24.
Here is the model in brms
m <- brm(abs ~ group*time + (1|id),
data = df,
family = 'bernoulli',
prior = set_prior("normal(0,5)"),
warmup = 1e3,
iter = 1.1e4,
chains = 4,
cores = 4,
seed = 1234)
print(m, pars = "b")
# output
# Family: bernoulli
# Links: mu = logit
# Formula: abs ~ group * time + (1 | id)
# Data: df (Number of observations: 440)
# Samples: 4 chains, each with iter = 11000; warmup = 1000; thin = 1;
# total post-warmup samples = 40000
#
# Group-Level Effects:
# ~id (Number of levels: 128)
# Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
# sd(Intercept) 4.12 0.84 2.73 6.02 1.00 12899 21612
#
# Population-Level Effects:
# Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
# Intercept -10.60 1.90 -14.67 -7.24 1.00 16235 21747
# grouptreatment -0.59 1.94 -4.49 3.13 1.00 19024 24606
# time4 4.93 1.53 2.17 8.15 1.00 21260 25054
# time8 6.60 1.57 3.76 9.90 1.00 20332 23917
# time12 6.35 1.59 3.46 9.69 1.00 21334 25006
# time24 5.71 1.56 2.85 8.98 1.00 22546 25676
# grouptreatment:time4 1.62 1.95 -2.10 5.53 1.00 21214 27577
# grouptreatment:time8 1.85 1.95 -1.90 5.77 1.00 20731 26607
# grouptreatment:time12 1.88 1.97 -1.89 5.87 1.00 20978 28585
# grouptreatment:time24 4.26 2.04 0.37 8.37 1.00 21738 28906
#
# Samples were drawn using sampling(NUTS). For each parameter, Eff.Sample
# is a crude measure of effective sample size, and Rhat is the potential
# scale reduction factor on split chains (at convergence, Rhat = 1).
Now, this model converges, unlike equivalent frequentist models in geepack, lme4, GLMMadaptive and glmmTMB packages. I assume this is because of the regularising priors helping guide the algorithm which values to spend its time exploring.
The thing that troubles me is the size of the log-odds coefficients for the grouptreatment:time24. Exponentiating 4.26 yields an odds ratio of 70.8! This does not seem like a reliable estimate to me, given that the proportion abstinent at this time point in the placebo group is 6/29 = 20.7% and the proportion in the treatment group is 14/26 = 53.8%
So why are these log-odds coefficients so high?
The output calls them ‘population-level’ effects but is it possible they in fact subject-specific effects of the sort usually returned by these models (see https://stats.stackexchange.com/questions/444380/confused-about-meaning-of-subject-specific-coefficients-in-a-binomial-generalise)?
There is a function in brms called ‘conditional_effects’. Would this provide better estimates of the odds ratio at week 24?
Another possibility that occurred to me is that the estimates at week 24 are unstable because there are so few data at this time point (n=55) compared to week 0 (n=128).
I guess my problem is I don’t run a whole lot of panel models with binary outcomes. Maybe the estimates of the week-24 group effect are fine, but my gut tells me they’re not. I just need some advice from people with more experience with these sorts of models.