Hi all,
As mentioned in the stan dev call I’ve been running SBC on the benchmark models:
I’m using cmdstan called via a python script which calculates the SBC histograms for a given model. I’m using cmdstan / python rather than something like cmdstanpy as my objective is to use the script to verify SMC-Stan, which currently exists as a cmdstan fork.
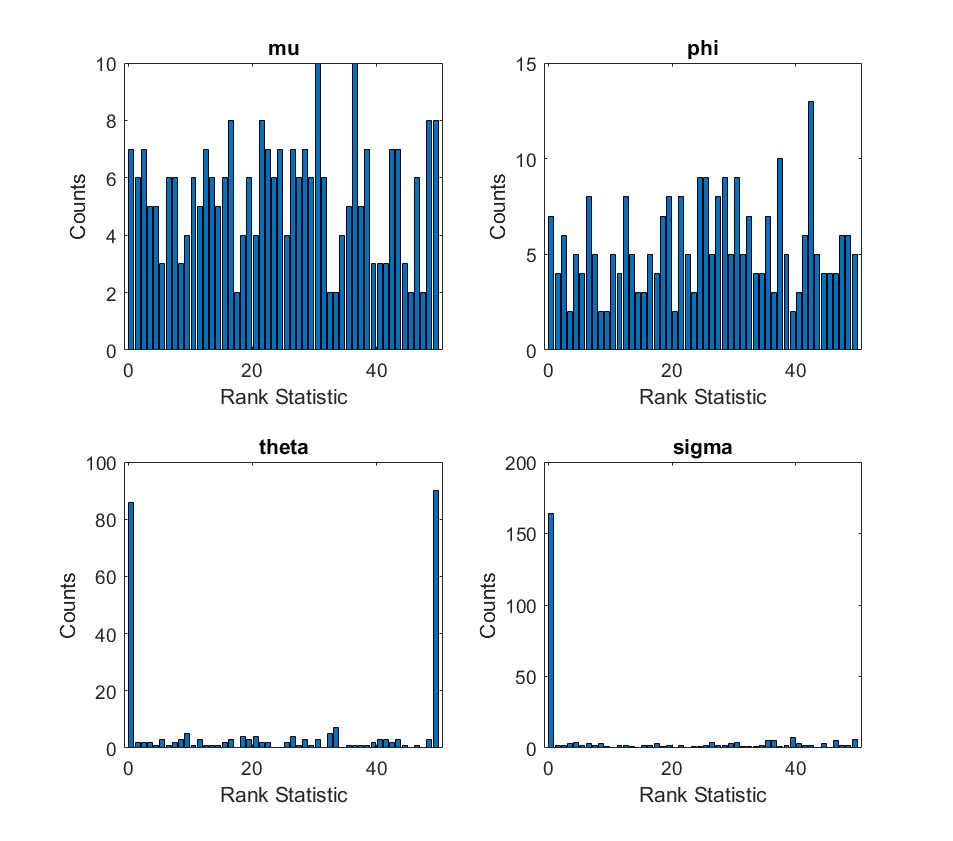
I managed to reproduce the results for the eight_schools model, but I’m seeing extremely skewed histograms for the arma model (shown below).
Here’s the model code I’m using:
// ARMA(1, 1)
functions {
real half_cauchy_rng(real lb) {
real p = cauchy_cdf(lb, 0, 2.5);
real u = uniform_rng(p, 1);
real y = 2.5*tan(pi()*(u - 0.5)); // inverse cauchy_cdf
return y;
}
}
data {
int<lower=1> T; // number of observations
real y[T]; // observed outputs
}
transformed data {
real y_sim[T];
real mu_sim;
real phi_sim;
real theta_sim;
real<lower=0> sigma_sim;
vector[T] nu_sim; // prediction for time t
vector[T] err_sim; // error for time t
mu_sim = normal_rng(0, 10);
phi_sim = normal_rng(0, 2);
theta_sim = normal_rng(0, 2);
sigma_sim = half_cauchy_rng(0);
for (t in 1:T){
err_sim[t] = normal_rng(0, sigma_sim);
}
nu_sim[1] = mu_sim + phi_sim * mu_sim; // assume err[0] == 0
y_sim[1] = err_sim[1] + nu_sim[1];
for (t in 2:T) {
nu_sim[t] = mu_sim + phi_sim * y_sim[t - 1] + theta_sim * err_sim[t - 1];
y_sim[t] = err_sim[t] + nu_sim[t];
}
}
parameters {
real mu; // mean coefficient
real phi; // autoregression coefficient
real theta; // moving average coefficient
real<lower=0> sigma; // noise scale
}
model {
vector[T] nu; // prediction for time t
vector[T] err; // error for time t
mu ~ normal(0, 10);
phi ~ normal(0, 2);
theta ~ normal(0, 2);
sigma ~ cauchy(0, 2.5);
nu[1] = mu + phi * mu; // assume err[0] == 0
err[1] = y[1] - nu[1];
for (t in 2:T) {
nu[t] = mu + phi * y[t - 1] + theta * err[t - 1];
err[t] = y[t] - nu[t];
}
err ~ normal(0, sigma);
}
generated quantities {
int<lower=0,upper=1> lt_sim[4];
lt_sim[1] = mu < mu_sim;
lt_sim[2] = phi < phi_sim;
lt_sim[3] = theta < theta_sim;
lt_sim[4] = sigma < sigma_sim;
}
Does anyone have ideas as to what might be going on?