I am doing a simulation study for my multiple quantile regression model that involves fitting a piece-wise likelihood. Following is my problem set up:
where 0=\kappa_0<\kappa_1<\dots<\kappa_{L-1}<\kappa_L=1 are breakpoints representing multiple quantiles, and
is the proposed nonlinear true quantile process. \alpha_l=\begin{pmatrix}\alpha_{0l}\\\alpha_{1l}\end{pmatrix} is a vector of two elements for every l.
In order to ensure the monotonicity of quantiles, I generate \alpha_l^* first and then apply restrictions to form \alpha_l. My priors are
and \alpha_l^* is further transformed to \alpha_l using a self-defined function.
Case 1: L=2, \kappa_1=0.5, f_0\sim \phi, i.e. a nonlinear median regression. The goal is to recover \alpha_l, l =1,2.
functions{
matrix thresh(matrix alpha_star, real eps){
int M = rows(alpha_star);
int N = cols(alpha_star);
matrix[M,N] alpha_out;
for(l in 1:N){
real WC = alpha_star[1,l]-sum(fabs(alpha_star[2:M,l]));
if(WC < eps){
alpha_out[1,l] = eps;
alpha_out[2:M,l] = rep_vector(0,M-1);
}else{
alpha_out[,l] = alpha_star[,l];
}
}
return alpha_out;
}
int which_less(row_vector q_vec, real a){
int loc = 1;
while((loc<=num_elements(q_vec))&&(q_vec[loc]<a)){
loc += 1;
}
return loc-1;
}
}
data {
int<lower=0> N;
int L;
vector[N] y;
matrix[N,2] x;
matrix[L,L-1] basis_m;
vector[L-1] q_base;
}
// The parameters accepted by the model. Our model
// accepts two parameters 'mu' and 'sigma'.
parameters {
vector[2] mu;
matrix[2,L] alpha_star;
vector<lower=0> [2] prec;
}
transformed parameters{
vector<lower=0> [2] sigma;
matrix[2,L] alpha;
for(j in 1:2){
sigma[j] = 1/sqrt(prec[j]);
}
alpha = thresh(alpha_star,0.1);
}
model {
matrix[N,L] x_a;
matrix[N,L-1] q_model;
mu ~ normal(0,10);
prec ~ gamma(0.3,0.3);
for(j in 1:2){
alpha_star[j,] ~ normal(mu[j],sigma[j]);
}
x_a = x*alpha;
q_model = x_a*basis_m;
for(i in 1:N){
if(y[i]<=q_model[i,1]){
y[i] ~ normal(0,x_a[i,1]) T[,q_model[i,1]];
}else{
if(y[i]>q_model[i,L-1]){
y[i] ~ normal(q_model[i,L-1]-x_a[i,L]*q_base[L-1],x_a[i,L]) T[q_model[i,L-1],];
}else{
int idx = which_less(q_model[i,],y[i]);
y[i] ~ normal(q_model[i,idx]-x_a[i,idx+1]*q_base[idx],x_a[i,idx+1]) T[q_model[i,idx],q_model[i,idx+1]];
}
}
}
}
I first generate a random design matrix and then generate true quantile process based on the design matrix. For this case the model did a perfect job recovering the parameters of interest.
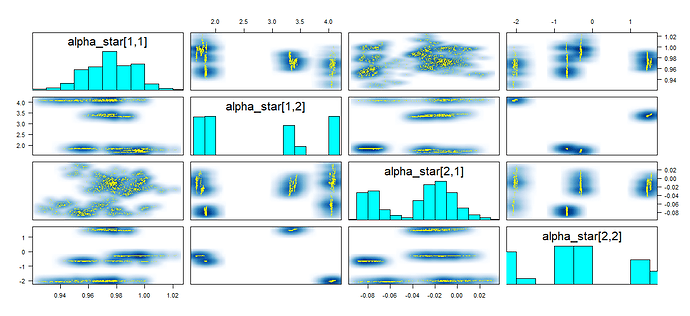
Case 2: L=4, \kappa_1=0.25,\kappa_2=0.5,\kappa_3=0.75,f_0\sim \phi, i.e. a nonlinear multiple quantile regression. The goal is again to recover \alpha_l, l =1,2,3,4. I used the same code and simulation procedure as listed above, but this time the results are really bad.
First of all, the sampling is really inefficient. With 1000 iterations, 250 warm-up it takes minutes whereas only seconds is needed in the first case. Secondly, nearly all transitions reach the default maximum tree depth (10) and the traceplot displays high autocorrelation. Lastly, when I attempt to increase the maximum tree depth from 10 to 30, the program just will not start or finish warm-up stage even after hours.
I have tried hours to detect and fix problems in my model and coding but there hadn’t been any improvement. Do you guys see any obvious problems from the model or code ? What are some guidelines to diagnose my model in this case. I really appreciate any help.