Hi
I’m learning how to fit survival analysis models in Stan. I’m using a dataset that Richard McElreath used in this lecture (minute 31:00): https://www.youtube.com/watch?v=p7g-CgGCS34&list=WL&index=19
This is a dataset of cat adoptions in an animal care facility. The goal is to determine if black cats are less likely to be adopted than other cats.
I’m attaching the dataset and sharing the code I used for this. I already had help from this forum here.
Data preparation:
d <- AustinCats
d$adopt <- ifelse( d$out_event=="Adoption" , 1L , 0L )
dat <- list(
N=nrow(d),
days_to_event = as.numeric( d$days_to_event ),
color_id = ifelse( d$color=="Black" , 1L , 2L ) ,
adopted = d$adopt
)
Stan model:
data{
int N;
int adopted[N];
vector[N] days_to_event;
int color_id[N];
}
parameters{
vector[2] a;
}
transformed parameters{
vector[N] lambda;
vector[N] mu;
for ( i in 1:N ) {
mu[i] = a[color_id[i]];
mu[i] = exp(mu[i]);
}
for ( i in 1:N ) {
lambda[i] = 1/mu[i];
}
}
model{
a ~ normal( 0 , 1 );
for ( i in 1:N )
if ( adopted[i] == 0 ) target += exponential_lccdf(days_to_event[i] | lambda[i]);
for ( i in 1:N )
if ( adopted[i] == 1 ) days_to_event[i] ~ exponential( lambda[i] );
}
generated quantities{
real pred[N];
for(i in 1:N){
pred[i]=exponential_rng(lambda[i]);
}
}
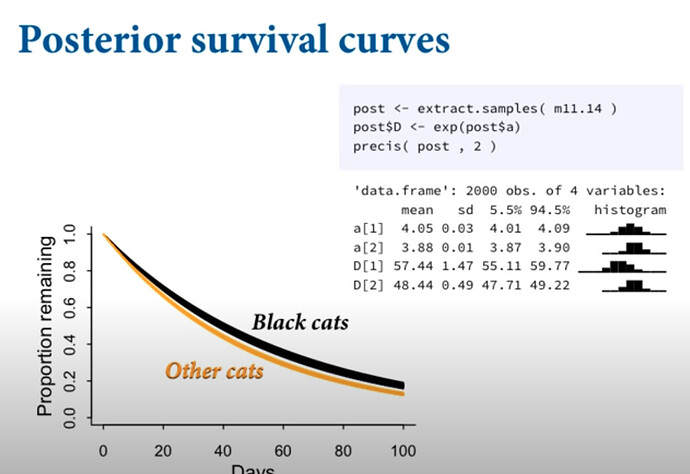
In the slides, Richard McElreath fitted an identical model and plotted it as folllows:
I would like to do two things:
-
learn how to make a similar plot
-
learn how to perform posterior predictive checks so that I can make sure the model fits the data
Can anyone share any hints on how I can do this? Or recommend any resources?
I would like to avoid brms and rstanarm for the time being. I’m trying to learn “pure” Stan.
Thanks
AustinCats.csv (2.8 MB)