Okay, so reporting back in with perhaps stronger evidence of something funky happening with the affine transform in this logit-normal hierarchical model. And while it does seem to have something to do with initial values, it doesn’t appear to be anything in particular about low initial values for sigma.
As suggested I am comparing two “identical” models fit to the same data with the same initial values and the same seed.
As @Bob_Carpenter suggested here are two separate scripts:
binomial_hier_affine.stan
data {
int<lower = 0> I;
array[I] int N, y;
}
parameters {
real mu;
real<lower = 0> sigma;
vector<offset=mu, multiplier=sigma>[I] logit_p;
}
model{
mu ~ student_t(3, 0, 2.5);
sigma ~ loglogistic(1, 4);
logit_p ~ normal(mu, sigma);
y ~ binomial_logit(N, logit_p);
}
and
binomial_hier_raw.stan
data {
int<lower = 0> I;
array[I] int N, y;
}
parameters {
real mu;
real<lower = 0> sigma;
vector[I] logit_p_raw;
}
transformed parameters{
vector[I] logit_p = mu + logit_p_raw * sigma;
}
model{
mu ~ student_t(3, 0, 2.5);
sigma ~ loglogistic(1, 4);
logit_p_raw ~ std_normal();
y ~ binomial_logit(N, logit_p);
}
In R we simulate overdispersed binomial data like so:
I = 500
p = plogis(rnorm(I, mean = 0.69, sd = 0.5))
N = rpois(I, rgamma(I, 20, 1))
y = rbinom(I, N, p)
x = rbinom(I, N, 0.69)
summary(glm(cbind(y, N-y) ~ NULL, family = "quasibinomial"))
We can see that the data are consistent with extrabinomial variation by checking with glm()
(Dispersion parameter for quasibinomial family taken to be 1.935123)
Here is function to generate initial values ensuring that the both models receive the same initial values for both sigma and logit_p/logit_p_raw parameters
init_binom <- function(mu_in, logit_p_raw_in, sigma_in, rand = T){
inits = list()
for (i in 1:length(sigma_in)){
if(rand){
logit_p_raw = runif(length(logit_p_raw_in[,1]), -2, 2)
sigma_in = exp(runif(1, -2, 2))
mu_in = runif(1, -2, 2)
inits[[i]] = list(mu = mu_in, sigma = sigma_in,
logit_p = mu_in + logit_p_raw*sigma_in,
logit_p_raw = logit_p_raw)
} else{
logit_p_raw = logit_p_raw_in
inits[[i]] = list(mu = mu_in[i], sigma = sigma_in[i],
logit_p = mu_in[i] + logit_p_raw[,i]*sigma_in[i],
logit_p_raw = logit_p_raw[,i])
}
}
return(inits)
}
And some initial values tests, one deliberately checking low initial values of sigma and holding other parameters fixed and the other checking with random values that I think should be similar to how Stan generates them.
mu_init = rep(0.69, 8)
sigma_chain = c(1e-21, 1e-15, 1e-10, 1e-6, 1e-3, 0.01, 0.1, 1)
logit_p_raw_init = matrix(nrow = I, ncol = 8)
for (i in 1:8)
logit_p_raw_init[,i] = rep((0.7 - mu_init[i]) / sigma_chain[i], I)
set.seed(125)
inits_nonrandom = init_binom(mu_init, logit_p_raw_init, sigma_chain, F)
inits = init_binom(mu_init, logit_p_raw_init, sigma_chain, T)
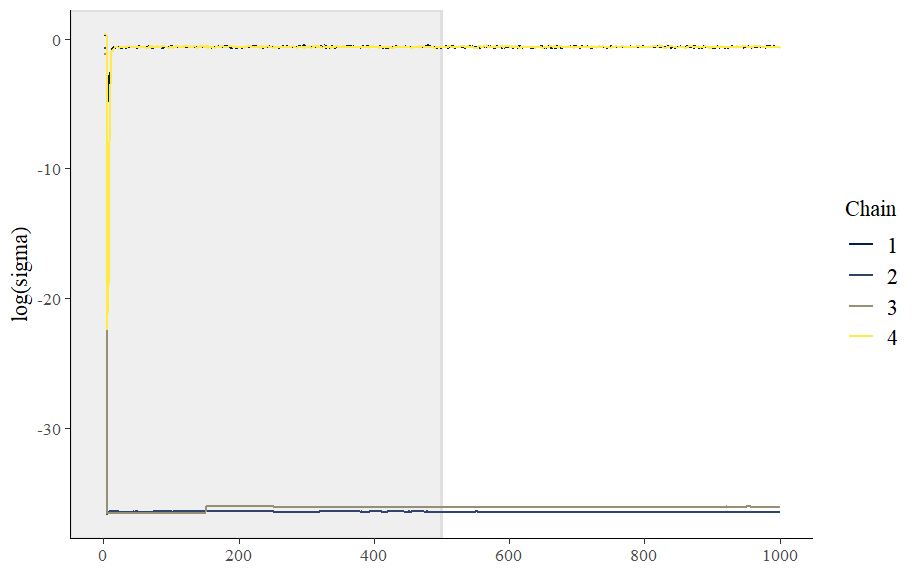
Running the models with non-random inits shows that the affine transform reliably fails when sigma is initialized to low values. The manual version also struggled (with one chain giving me treedepth warnings) but all of the chains converged.
First a check that the models are initializing to the same values as suggested earlier in this thread:
init_test_affine = binom_hier_fit_affine$draws(variables = c("mu", "sigma", "logit_p[1]", "lp__"), inc_warmup = T)
init_test_raw = binom_hier_fit_raw$draws(variables = c("mu", "sigma", "logit_p[1]", "lp__"), inc_warmup = T)
> init_test_affine[1:6,,"sigma"]
# A draws_array: 6 iterations, 8 chains, and 1 variables
, , variable = sigma
chain
iteration 1 2 3 4 5 6 7 8
1 1e-21 4.7e-90 4.4e-159 1.0e-06 0.1210 0.26 0.34 0.80
2 1e-21 4.7e-90 4.4e-159 1.0e-06 0.1210 0.26 0.34 0.80
3 1e-21 3.6e-67 4.4e-159 6.7e-04 0.1210 0.26 0.34 0.80
4 1e-21 1.3e-63 4.7e-159 2.3e-264 0.1210 0.26 0.34 0.80
5 1e-21 1.3e-63 6.6e-159 2.2e-264 0.0026 0.38 0.45 0.74
> init_test_raw[1:5,,"sigma"]
# A draws_array: 5 iterations, 8 chains, and 1 variables
, , variable = sigma
chain
iteration 1 2 3 4 5 6 7 8
1 1.0e-21 2.8e-13 1.0e-10 1.0e-06 0.1210 0.26 0.34 0.80
2 1.0e-21 2.8e-13 1.0e-10 1.0e-06 0.1210 0.26 0.34 0.80
3 1.5e-21 2.8e-13 9.4e-09 6.7e-04 0.1210 0.26 0.34 0.80
4 1.7e-20 9.5e-242 8.5e-153 2.3e-264 0.1210 0.26 0.34 0.80
5 2.5e-45 1.2e-241 1.1e-152 3.1e-264 0.0026 0.38 0.45 0.74
> init_test_affine[1:5,,"logit_p[1]"]
# A draws_array: 5 iterations, 8 chains, and 1 variables
, , variable = logit_p[1]
chain
iteration 1 2 3 4 5 6 7 8
1 0.71 31233 4097 0.7 0.44 0.64 0.55 1.3
2 0.71 31233 4097 0.7 0.44 0.64 0.55 1.3
3 0.71 -9704 4097 330.3 0.44 0.64 0.55 1.3
4 0.71 3404 3699 225.2 0.44 0.64 0.55 1.3
5 0.71 3404 3524 -18.9 0.64 0.97 1.04 1.7
> init_test_raw[1:5,,"logit_p[1]"]
# A draws_array: 5 iterations, 8 chains, and 1 variables
, , variable = logit_p[1]
chain
iteration 1 2 3 4 5 6 7 8
1 0.7 -322 0.7 0.7 0.44 0.64 0.55 1.3
2 0.7 -322 0.7 0.7 0.44 0.64 0.55 1.3
3 -3.7 -322 325.2 330.3 0.44 0.64 0.55 1.3
4 176.4 -133 125.5 225.2 0.44 0.64 0.55 1.3
5 492.3 209 -56.6 43.0 0.64 0.97 1.04 1.7
That’s a qualified yes, though I’m not sure what’s going on with chains 2 and 3.
Traceplots. For the affine transform model, all four chains with sigma initialized < 1e-5 got stuck with sigma near-zero, but oddly they all bottomed out far below the initialized values before somewhat recovering.
Here are the traces for manual non-centering model:
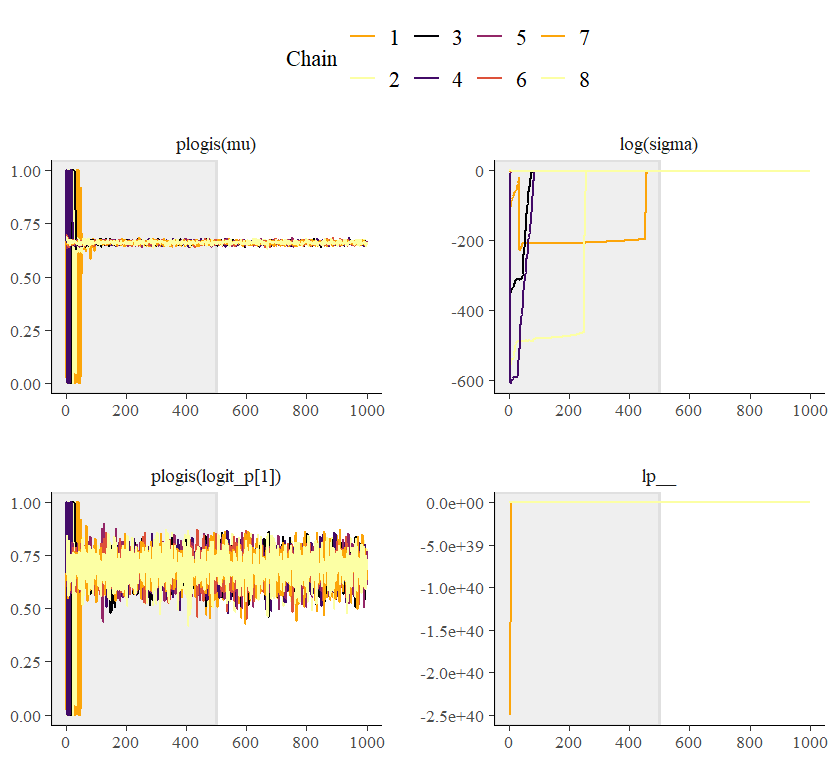
Now for something very odd with the the random inits. I was able to pretty quickly hit upon a combination that resulted in a single chain getting stuck at zero for sigma using the affine transform version:
But the initial value for chain 4 was pretty high for sigma. The value provided was 2.26, which got turned into 0.84 for the first few warm-up iterations:
> unlist(c(inits[[1]][2], inits[[2]][2], inits[[3]][2], inits[[4]][2],
+ inits[[5]][2], inits[[6]][2], inits[[7]][2], inits[[8]][2]))
sigma sigma sigma sigma sigma sigma sigma sigma
0.2335458 6.3053346 4.5656075 2.2926901 0.2683244 0.4980496 0.2839859 4.6789906
> init_test_affine[1:5,,"sigma"]
# A draws_array: 5 iterations, 8 chains, and 1 variables
, , variable = sigma
chain
iteration 1 2 3 4 5 6 7 8
1 2.3e-01 2.42 0.026 0.84 2.3e-13 0.43 0.14313 2.8e-01
2 2.3e-01 2.42 0.026 0.84 2.3e-13 0.43 0.14313 2.8e-01
3 2.3e-01 2.42 0.026 0.84 2.3e-13 0.43 0.14313 2.8e-01
4 2.3e-01 2.42 0.026 0.84 2.3e-13 0.43 0.14313 2.8e-01
5 8.0e-08 0.38 0.148 0.39 8.8e-02 0.29 0.00021 1.5e-15
> init_test_raw[1:5,,"sigma"]
# A draws_array: 5 iterations, 8 chains, and 1 variables
, , variable = sigma
chain
iteration 1 2 3 4 5 6 7 8
1 2.3e-01 2.42 0.026 0.84 2.3e-13 0.43 0.14313 2.8e-01
2 2.3e-01 2.42 0.026 0.84 2.3e-13 0.43 0.14313 2.8e-01
3 2.3e-01 2.42 0.026 0.84 2.3e-13 0.43 0.14313 2.8e-01
4 2.3e-01 2.42 0.026 0.84 2.3e-13 0.43 0.14313 2.8e-01
5 8.0e-08 0.38 0.148 0.39 8.8e-02 0.29 0.00021 4.0e-15
Plotting the paths of the two models for the first 20 iterations (iteration number labelled) and you can see them track each other pretty well at first. But then chain 4 gets stuck for the affine transform. The other two chains (5 and 8) are displayed because for these chains values also started to get down near zero for sigma but then recovered (none of the other chains ever got quite so low). So I’m really not clear what combination of conditions is resulting in the affine transform performing so poorly with this model.
In summary, there is something different about the affine transformation compared to the manual non-centered transform that results in different behavior. Identical initial values and seeds will diverge and it’s not solely a feature of poor initial conditions in terms of sigma being set to zero. However, what’s going on here is a sort of triple transformation (lower bound for sigma, affine/non-centering for the logit_p’s, and then p for binomial. So maybe something pathological in those combinations of constraints?