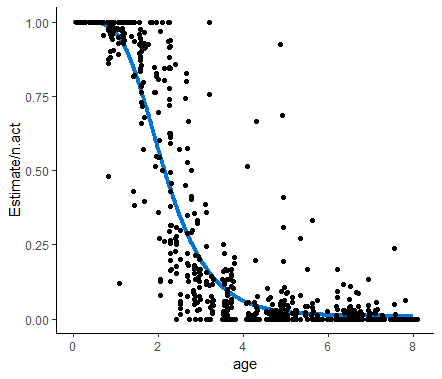
Hi all, I am trying to fit a non-linear mixed effects model to a maternal behaviour. The response variable is a discontinuous proportion. Therefore, it is in the form of a matrix with two columns (resp.mat): 1. no. of successes 2. no. of trials. I want to model this matrix as a function of offspring age and other predictors. The other predictors include offspring sex with two levels, parity with two levels, male status with two levels, and two other continuous predictors. I have one random factor: mother ID. Offspring age has a non-linear effect. Gompertz function fits the non-linear effect well (graph included). I want to add followID as a random factor so that the successes and trials within one follow is treated as a single data point and not multiple data points (which would result in pseudo replication). I have no prior experience with fitting a non-linear mixed effects model. So, I would like some help to understand how to specify the model in brms once I have a function for offspring age, especially with respect to fixed and random effect specification. I am pasting the code I have so far.
gompertz.pred.fun ← function(x, pars){
a = as.vector(pars[“a”])
b = as.vector(pars[“b”])
cc = as.vector(pars[“cc”])
return(1-plogis(a)*exp(-exp(b)*exp(-exp(cc)*x)))
}
gompertz.LL.fun ← function(y, pred.y){
return(-sum(dbinom(x = y[, 1], size=apply(X = y, MARGIN = 1, FUN = sum), prob = pred.y, log = T)))
}
gompertz.both.fun ← function(pars, x, y){
pred.y = gompertz.pred.fun(x = x, pars = pars)
gompertz.LL.fun(y = y, pred.y = pred.y)
}
x = seq(from = 0, to = max(xdata$offspringAge), length.out = 100)
pred.y = gompertz.pred.fun(x = x, pars = c(a = 10, b = 1, cc = 1))
plot(x, pred.y, type = “l”)
gompertz.both.fun(x = xdata$offspringAge, y = xdata$resp.mat, pars = c(a = 10, b = 1, cc = 1))
res = optim(par = c(a = 10, b = 1, cc = 1), fn = gompertz.both.fun, x = xdata$offspringAge, y = xdata$resp.mat)
pred.y = gompertz.pred.fun(x = x, pars = res$par)
plot(x = xdata$offspringAge, y = xdata$resp.mat[, 1]/apply(X = xdata$resp.mat, MARGIN = 1, FUN = sum))
lines(x=x, y=pred.y, col=“red”)
xdata$n.act = apply(X = xdata$resp.mat, MARGIN = 1, FUN = sum)
xdata$n = xdata$resp.mat[, 1]
brm.formula = brmsformula(formula = n|trials(n.act) ~ 1-exp(a)/(1+exp(a))*exp(-exp(b)*exp(-exp(cc)*z.age)), #(1|followID)?
flist = NULL, family = binomial, nl = T, #autocorr=NULL,
a~1+(1|motherID),
b~1+(1|motherID),
cc~1+(1|motherID)
)
get_prior(brm.formula, data = xdata)
priors=c(
set_prior(“normal(4.5, 3)”, class=“b”, nlpar=“a”),
set_prior(“normal(2.5, 3)”, class=“b”, nlpar=“b”),
set_prior(“normal(0.3, 3)”, class=“b”, nlpar=“cc”),
set_prior(“exponential(1)”, class=“sd”, nlpar=“a”, group=“offspringID”),
set_prior(“exponential(1)”, class=“sd”, nlpar=“b”, group=“offspringID”),
set_prior(“exponential(1)”, class=“sd”, nlpar=“cc”, group=“offspringID”),
set_prior(“exponential(1)”, class=“sd”, nlpar=“a”, group=“motherID”),
set_prior(“exponential(1)”, class=“sd”, nlpar=“b”, group=“motherID”),
set_prior(“exponential(1)”, class=“sd”, nlpar=“cc”, group=“motherID”))
Where should priors for followID be included?
Here is the Gompertz function fit to to the data: