Hi everyone, how are you guys doing.
I am new to Rstan. I wrote a Rstan code to sample from proportional hazards models. I tried simulation data of different sample sizes and random seeds. It gives me errors stopping the calculation for as high as 20% of the simulations at particular settings.
The messages are as follows:
"
DIAGNOSTIC(S) FROM PARSER:
Info: left-hand side variable (name=betax) occurs on right-hand side of assignment, causing inefficient deep copy to avoid aliasing.
No C++ compiler found, so the following will probably not work.
See https://github.com/stan-dev/rstan/wiki/Installing-RStan-on-Windows#toolchain
Warning message:
In system(cmd) : ‘make’ not found
ERROR(s) during compilation: source code errors or compiler configuration errors!
Program source: (it is too long, I cant paste it here)
Error in compileCode(f, code, language = language, verbose = verbose) :
Compilation ERROR, function(s)/method(s) not created! Warning message:
In system(cmd) : ‘make’ not found
In addition: Warning messages:
1: In fitter(X, Y, strats, offset, init, control, weights = weights, :
Loglik converged before variable 6 ; beta may be infinite.
2: In system2(file.path(Sys.getenv(“R_HOME”), “bin”, Sys.getenv(“R_ARCH_BIN”), :
running command ‘“C:/PROGRA~1/R/R-35~1.1/bin//R” CMD config CXX’ had status 1
3: In stan_model(file, model_name = model_name, model_code = model_code, :
StanHeaders version is ahead of rstan version; update to latest rstan
4: In system(cmd, intern = !verbose) :
running command ‘C:/PROGRA~1/R/R-35~1.1/bin/x64/R CMD SHLIB file34e43ec72e29.cpp 2> file34e43ec72e29.cpp.err.txt’ had status 1
"
Does anyone has the same issues? May I get a hint? Thank you very much in advance.
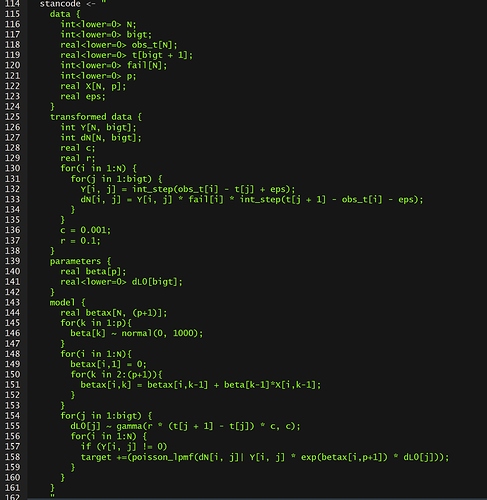
Here is my code:
stan(
model_code = stancode, # Stan program
data = dat, # named list of data
chains = 1, # number of Markov chains
warmup = burn, # number of warmup iterations per chain
iter = nsample+burn # total number of iterations per chain
)
It didn’t compile, so I don’t know what is the 20% you are referring to. What happens when you do
pkgbuild::has_build_tools(debug = TRUE)
?
1 Like
Hi Bgoodri, thank you for your reply.
I am sorry I did not make it clear. I usually run hundreds of simulations with set seeds. For example, 500 simulated sets of data, should give me 500 results. But due to errors, I only got about 400 in rare occasions. Most cases the error rate is less than 3%.
The command returns a FALSE. I installed the package again, still gets errors :(
That is what I am not understanding. If pkgbuild::has_build_tools(debug = TRUE) returns FALSE, it should not even compile, in which case how does it run at all?
1 Like
Thank you for the info. I run the debug codes on my local . I made a mistake that I recalled the environment are the same, but actually they are not. I still haven’t figured it out why I got the error messages on my computer after install the package. But that can wait.
My main problem is on HPC. I just check the log, it seems the only error I have on HPC is this :
“recompiling to avoid crashing R session
Error in sink(type = “output”) : invalid connection”
I googled it, it seems like a file writing issue. It is probably not Stan’s fault :)
If something does not compile, you want to pass verbose = TRUE to stan or stan_model to see the entire error message and then post the part that includes the string error: (with the colon) here.
But in a HPC environment, you should not be recompiling models anyway. Compile it once from the login node after specifying rstan_options(auto_write = TRUE) and then the execute nodes should be reading the compiled version of the disk without recompiling.
1 Like
Thank you for the info. I am still testing to see what cause the connection problem. I will add verbose = TRUE to see what other error message will I get.