The aim of my model is to estimate the prevalence (e.g. of SARS-CoV-2 infection) over time from testing data, adjusting for test sensitivity and specificity. This is indeed a simplified version of my real model, but it is still able to reflect the issue I’m facing.
Let n_i the number of tests performed at week N (i = 1,\dots,N) and y_i the number of tests that are positive. A simple estimated of the prevalence would be y_i/n_i but this does not adjust for imperfect test sensitivity and specificity. We thus followed the method shown by Gelman et al., which takes into account the uncertainy about SARS-CoV-2 PCR test specificity and specificity by simultaneously fitting some specificity/sensitvity data together with the rest of the data.
Using R (version 4.1.3) and CmdStan (version 2.29), I simulated different datasets of number of total and positive tests (n and y) and applied Gelman’s method to estimate the true prevalence. The specificity data consists of 13 studies with an average specificity of 99.5% and the sensitivity data consists of 3 studies showing an average sensitivity of 82.8%.
As a first try, I varied n at around 200. I fixed N=50, with a prevalence varying over time as shown by the dots in the figure below. The model estimates well the observed prevalence (in red), with expected credibility intervals.

We also observe that the estimates of the sensitivity and specificity are not modified by the inference, which is what we expect and want, as the test results here (n and y) do not bring any new information on test specificity or sensitivity.
variable mean median sd mad q5 q95 rhat ess_bulk ess_tail
<chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 sens 0.810 0.812 0.0327 0.0331 0.753 0.861 1.00 912. 1395.
2 spec 0.995 0.995 0.00136 0.00134 0.992 0.997 1.00 5054. 2303.
Stan model
data {
//data to fit
int N;
array[N] int n; //number of pools
array[N] int y; //number of positive pools
//hyperparameters of the priors
array[2] real p_prev;
array[2] real p_sens;
array[2] real p_spec;
//data sensitivity specificity
int<lower = 0> J_spec;
array[J_spec] int <lower = 0> y_spec;
array[J_spec] int <lower = 0> n_spec;
int<lower = 0> J_sens;
array[J_sens] int <lower = 0> y_sens;
array[J_sens] int <lower = 0> n_sens;
int inference;
}
parameters {
//True prevalence
array[N] real <lower=0,upper=1> prev;
real <lower=0,upper=1> sens; //sensitivity
real <lower=0,upper=1> spec; //specificity
}
transformed parameters {
//Observed prevalence
array[N] real <lower=0,upper=1> obs_prev;
for(i in 1:N){
obs_prev[i] = prev[i] * sens + (1.0-prev[i]) * (1.0-spec);
}
}
model {
//priors
prev ~ beta(p_prev[1], p_prev[2]);
sens ~ beta(p_sens[1], p_sens[2]);
spec ~ beta(p_spec[1], p_spec[2]);
if(inference==1){
for(i in 1:N){
target += binomial_lpmf(y[i] | n[i], obs_prev[i]);
}
}
target += binomial_lpmf(y_sens | n_sens, sens);
target += binomial_lpmf(y_spec | n_spec, spec);
}
R script
library(cmdstanr)
library(ggplot)
library(dplyr)
mod = cmdstan_model(stan_file = "mod_example.stan")
#data
time=1:50
prev = 0.1 + 0.6 *time/50 - 0.6*(time/50)^2
plot(time,prev)
n = round(rnorm(50,20000,2000))
stan_data = list(
N = 50, #number of observations
n = n, #number of pools
y = rbinom(50,n,prev), #number of positive pools
p_prev = c(0.25,1),
J_spec = 13,
y_spec = c(368,30,70,1102,300,311,500,198,99,29,146,105,50),
n_spec = c(371,30,70,1102,300,311,500,200,99,31,150,108,52),
J_sens = 3,
y_sens = c(78,27,25),
n_sens = c(85,37,35),
p_sens = c(1,1),
p_spec = c(1,1),
inference=0)
# prior
samples_mod = mod$sample(data = stan_data,
chains = 4, parallel_chains = 4,adapt_delta=0.99,
iter_warmup = 1000, iter_sampling = 1000)
#results
samples_mod$summary(c("sens","spec"))
samples_mod$summary(c("prev","obs_prev")) %>%
tidyr::separate(variable,"\\[|\\]",into=c("variable","time","null")) %>%
mutate(time=as.numeric(time)) %>%
dplyr::select(time,variable,median,q5,q95) %>%
ggplot(aes(x=time)) +
geom_ribbon(aes(ymin=q5,ymax=q95,fill=variable),alpha=.1) +
geom_line(aes(y=median,colour=variable),size=1)+
geom_point(data=data.frame(time=1:50, prev),aes(x=time,y=prev))
# posterior
stan_data$inference=1
samples_mod = mod$sample(data = stan_data,
chains = 4, parallel_chains = 4,adapt_delta=0.99,
iter_warmup = 1000, iter_sampling = 1000)
#results
samples_mod$summary(c("sens","spec"))
samples_mod$summary(c("prev","obs_prev")) %>%
tidyr::separate(variable,"\\[|\\]",into=c("variable","time","null")) %>%
mutate(time=as.numeric(time)) %>%
dplyr::select(time,variable,median,q5,q95) %>%
ggplot(aes(x=time)) +
geom_ribbon(aes(ymin=q5,ymax=q95,fill=variable),alpha=.1) +
geom_line(aes(y=median,colour=variable),size=1)+
geom_point(data=data.frame(time=1:50, prev),aes(x=time,y=prev))
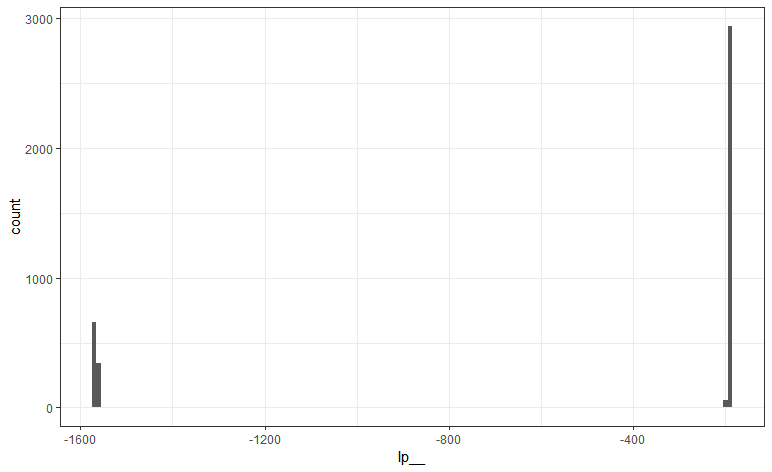
However, when I increase the number of tests n and varied them at around 20,000,
n = round(rnorm(50,20000,2000))
the model gives some weird results, with unexpected estimates of the true prevalence.

When looking at the sensitivity/specificity estimates, we see that the sensitivity is much lower than it should be:
variable mean median sd mad q5 q95 rhat ess_bulk ess_tail
<chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 sens 0.104 0.104 0.00215 0.00211 0.100 0.108 1.00 879. 1557.
2 spec 0.758 0.758 0.000965 0.000961 0.756 0.759 1.00 1207. 2402.
To me, it seems that fitting test data with high n gives too much important to this part of the likelihood and thus, makes the model able to adapt/update the sensitivity estimate. This should not be possible as, as said above, the testing data do not provide any new information about this estimate.
An idea to deal with this kind of issue could be to use a weighted likelihood. I went through this paper, but I’m not sure whether it is quite adapted to my problem and in addition, I do not see how to determine the weights beside the simple binomial example they showed.
Any thought?
Many thanks in advance!
Anthony