Hi
Thank you for the feedback!
For my richest dataset I have access to 608 survival data associated to 6919 observations that are nested in 1401 lesions that are nested in 1026 organs that are themselves nested in 608 patients.
And the model has 40 parameters if I’m only accounting for the population parameters. If I count each individual and lesion random effects, then I have 6065 parameters.
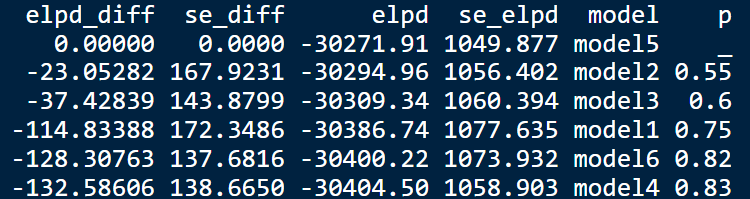
Grouping the data by patient, I chose to stick with the 5-fold cross validation (and not the 10 fold) due to computationnal cost. However, when I look at the elpd and se_elpd value of the 6 models that I compared, I feel like I’m saying that they are kinda equivalent.
(I also use the “p” added with Revised Uncertainty in Bayesian leave-one-out cross-validation based model comparison - #2 by avehtari)
Am I right? Is there something else that I could tried to see which model I should choose? I feel like the se_elpd are quite high? But I’m not sure if I can do something about it?
(I’ve tried using PSIS-LOO as leave-one-patient out and leave-one-measurement out, but my pareto k are too high (about 40% and 20% respectively), and using matching did not resolve the issue)
Thank you
-S