I want to fit a panel MSVAR model through hierarchical prior setting.
I built my DGP for 3 regimes, 2 lags,
and I simulate the data with 3 units 2 variables and 20 time length(I know this is quite small).
I sample it by using warmup=2000, post iteration=1000, with 4 chains
The illustrate expression is like: y_{t} \sim A * y_{t-p} + (\mu_{1} \quad \text{or} \quad \mu_{2} \quad \text{or} \quad \mu_{3}),
, where A is the VAR part regime-invariate coefficients; “\mu_{m}”, m=1, 2, 3, are the mean of each state.
and the prior is: A, \mu_{m} \sim N(\mu_{A}/\mu_{mm}, 1), \mu_{A}/\mu_{mm} \sim N(0, 10), Covariance follows wishart.
I found that when
adapt delta=0.8, the highest Rhat of A is likely 1.003, lp__ is 1.01;
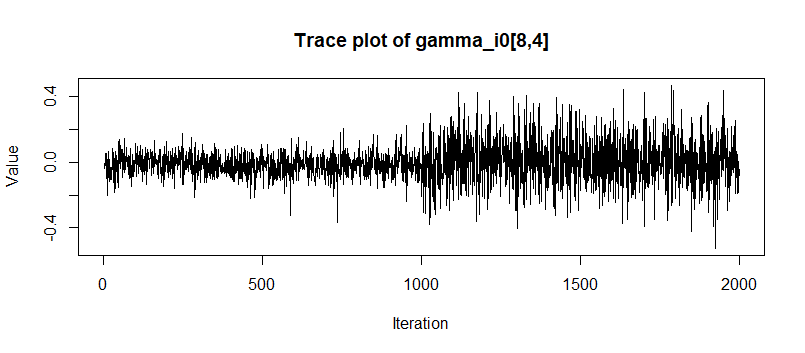
however, when I raise adapt delta to be higher, such as 0.99, the highest Rhat of A become 1.2, and the posterior draws of this coefficient will be like
or multimodality.
I’m wondering that it seems that in the previous results and article, as long as we give enough max_treedeepth and warmup, higher adapt delta will likely result better convergence. But in my DGP, the best adapt delta is at the middle b/w 0.8-0.85, higher will result multimodality(I confirm it with up to 4 chains to avoid random seed luck; I also set max_treedepth=15 and there was no reporting of hitting), and lower will result divergent. I feel it is strange that if I give enough space of max_treedepth to traversal the likelihood(higher adapt delta and higher treedepth), it should always give the better result.
I am not sure that my DGP scenario is because of 1) low sample size, and it may be too small so that multiple solutions of VAR coefs have the same likelihood, similar to label switching. 2) NUTS’s feature? whose suitable adapt delta may indeed can be concave. 3) auto adjusted step size of NUTS is not that smart, 4) or something else?