sircode_v2=’
functions {
real[] sir(real t, real[] y, real[] theta,
real[] x_r, int[] x_i) {
real S = y[1];
real I = y[2];
real R = y[3];
real N = x_i[1];
real beta = theta[1];
real gamma = theta[2];
real dS_dt = -beta * I * S / N;
real dI_dt = beta * I * S / N - gamma * I;
real dR_dt = gamma * I;
return {dS_dt, dI_dt, dR_dt};
}
}
data {
int<lower=1> n_days;
real y0[3];
real t0;
real ts[n_days];
int N;
int cases[n_days];
}
transformed data {
real x_r[0];
int x_i[1] = { N };
}
parameters {
real<lower=0> gamma;
real<lower=0> beta;
real<lower=0> phi_inv;
}
transformed parameters{
real y[n_days, 3];
real phi = 1. / phi_inv;
{
real theta[2];
theta[1] = beta;
theta[2] = gamma;
y = integrate_ode_rk45(sir, y0, t0, ts, theta, x_r, x_i);
}
}
model {
//priors
beta ~ normal(2, 1);
gamma ~ normal(0.4, 0.5);
phi_inv ~ exponential(5);
//sampling distribution
//col(matrix x, int n) - The n-th column of matrix x. Here the number of infected people
cases ~ neg_binomial_2(col(to_matrix(y), 2), phi);
}
generated quantities {
real R0 = beta / gamma;
real recovery_time = 1 / gamma;
real pred_cases[n_days];
pred_cases = neg_binomial_2_rng(col(to_matrix(y), 2), phi);
}
’
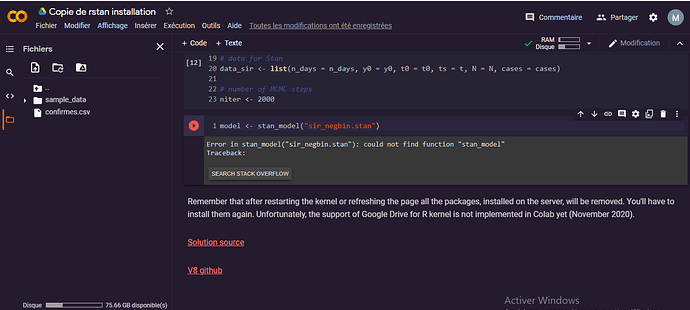
fit model
mod_v2 = stan_model(model_name = ‘sir_negbin.stan’,model_code = sircode_v2)
model <- stan_model(“sir_negbin.stan”)