Sure, happy to.
In short, some colleagues are planning a Registered Report that is similar in design to a study by Wood et al (2016) [https://link.springer.com/article/10.3758/s13423-015-0974-5\]
I can send you the data, or you can get it from ther OSF link.
My aim at present it to settle on an analysis pipeline. We will then do a simulation based justification of sample size to determine how many participants are required. My co-authors tell me they expect their effect to be larger than the one reported in this paper.
Here’s some initial data processing and summary plots:
library(tidyverse)
library(brms)
library(tidybayes)
library(patchwork)
options(mc.cores = 4)
d ← read_delim(“osf_wood2016/emotionXABraw_clean.txt”)
d %>% select(participant, condition = “Condition”,loc = “XABlocation”,correct = “XABresponse_2.corr”) %>% mutate(condition = factor(condition, labels = c(“control”, “gel”)),loc = (loc - 50)/50) %>%filter(is.finite(participant)) → d
d %>% group_by(condition, loc) %>%summarise(correct = mean(correct)) %>%ggplot(aes(loc, correct, colour = condition)) +geom_path() +geom_point()
d %>% group_by(participant, condition, loc) %>%summarise(correct = mean(correct)) → dagg
dagg %>% ggplot(aes(loc, correct, colour = condition)) +geom_smooth(method = lm,formula = y ~ x + I(x^2),color = “black”, linewidth = .5) +geom_point(size = 1) +facet_wrap(~participant) +theme_bw() +theme(panel.grid = element_blank(),strip.text = element_blank())
My colleagues are only interested in the difference between the two conditions. The loc variable is related to some face-morphing algorithm that was used to generate their stimuli. In the planned study, it is not of any special interest.
I can also do a plot per person and we can see there is quite a bit of variation underneath the grop averages:
Here is my attempt at modelling the data. (Note: the original paper used a different model, and attempts to account for the effect of loc by measuring the absolute value from each participant’s peak response. I’m not really sold on this approach, so have looked into using a quadratic or cubic)
full_priors ← c(prior(normal(0, 1.5), class = b, coef = “conditioncontrol”),prior(normal(0, 1.5), class = b, coef = “conditiongel”),
prior(normal(0, 1.5), class = b, coef = “conditioncontrol:loc”),
prior(normal(0, 1.5), class = b, coef = “conditiongel:loc”),
prior(normal(0, 1.5), class = b, coef = “conditioncontrol:IlocE2”),
prior(normal(0, 1.5), class = b, coef = “conditiongel:IlocE2”),
prior(exponential(5), class = sd))
m_full ← brm(correct ~ 0 + condition + condition:loc + condition:I(loc^2) +
(loc + I(loc^2) | participant),
data = d,
prior = full_priors,
family = “bernoulli”)`
summary(m_full)
Family: bernoulliLinks: mu = logitFormula: correct ~ 0 + condition + condition:loc + condition:I(loc^2) + (loc + I(loc^2) | participant)
Data: d (Number of observations: 21680)
Draws: 4 chains, each with iter = 2000; warmup = 1000; thin = 1;total post-warmup draws = 4000
Multilevel Hyperparameters:~participant (Number of levels: 121)
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
sd(Intercept) 0.57 0.05 0.48 0.67 1.00 1302 1865
sd(loc) 0.22 0.06 0.09 0.33 1.01 809 578
sd(IlocE2) 0.39 0.11 0.18 0.60 1.01 963 1904
cor(Intercept,loc) -0.01 0.19 -0.36 0.37 1.00 2421 1596
cor(Intercept,IlocE2) -0.77 0.13 -0.97 -0.48 1.00 1507 1844
cor(loc,IlocE2) 0.24 0.30 -0.41 0.75 1.00 1040 1262
Regression Coefficients:Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
conditioncontrol 1.64 0.08 1.48 1.81 1.00 1135 1999
conditiongel 1.34 0.08 1.19 1.50 1.00 961 1883
conditioncontrol:loc 0.21 0.05 0.10 0.32 1.00 4475 3075
conditiongel:loc 0.14 0.05 0.04 0.24 1.00 4385 3109
conditioncontrol:IlocE2 -1.07 0.12 -1.30 -0.84 1.00 3042 2734
conditiongel:IlocE2 -0.90 0.10 -1.10 -0.70 1.00 2256 2955
Draws were sampled using sampling(NUTS). For each parameter, Bulk_ESSand Tail_ESS are effective sample size measures, and Rhat is the potentialscale reduction factor on split chains (at convergence, Rhat = 1).
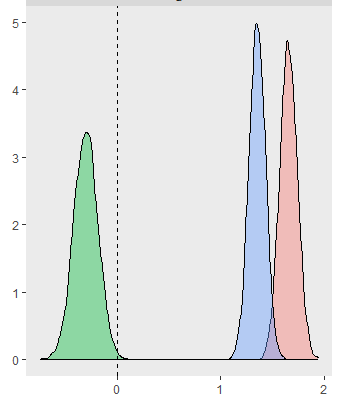
I get reasonable fits to the individual participant’s performance:
Posterior densities (and difference between…)
And, some more plots:
Finally, some model comparison:
priors <- c(prior(normal(0, 1.5), class = Intercept),
prior(normal(0, 1.5), class = b),
prior(exponential(5), class = sd))
m_null <- brm(correct ~ loc + I(loc^2) +
(loc + I(loc^2) |participant),
data = d,
prior = priors,
family = "bernoulli")
model_weights(m_full, m_null)
m_full m_null
0.0001219223 0.9998780777
We get some low ESS warnings here, but I’m not too worried about this at present. Maybe I am wrong about this, but I’d rather work out what I want to be going, before polishing things.
EDIT by Aki: fixed summary output formatting