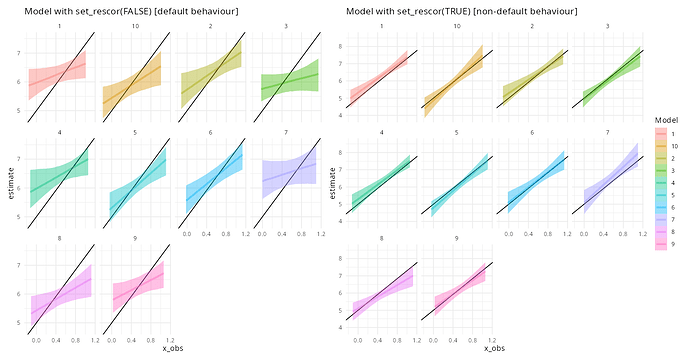
When both x and y are repeatedly measured and there is a group-level relationship between them and there is missing data, the default for brms results in a misspecified model (I think!).
make_data <- function(
n_groups=20,
n_per_group = 5,
y_int_mean = 5,
y_int_sd=.5,
y_sigma = 0.35,
x_coef=2.3,
x_sigma=0.1,
seed=1,
miss_type = c("random","groupwise"),
missingness = 0,
output_type=c("full","pergroup"),
plot=FALSE
){
miss_type <- match.arg(miss_type,miss_type)
output_type <- match.arg(output_type,output_type)
set.seed(seed)
n_groups <- 20
y_int_mean <- 5
y_int_sd <- 0.5
y_int_groups <- rnorm(n_groups,0,y_int_sd)
x_coef <- 2.3
x_true <- runif(n_groups)
y_true <- y_int_mean + y_int_groups + x_true*x_coef
# Take repeated measurements of x_true and y
n_per_group <- 5
y_sigma <- 0.35
y_obs <- unlist(sapply(y_true,\(i)rnorm(n_per_group,i,y_sigma),simplify = F))
x_sigma <- 0.1
x_obs <- unlist(sapply(x_true,\(i)rnorm(n_per_group,i,x_sigma),simplify = F))
groups <- rep(1:n_groups,each=n_per_group)
if(plot){
plot(y_obs~x_obs,col=groups)
abline(lm(y_obs~x_obs),lty=2)
abline(a=y_int_mean,b=x_coef)
legend(
"topleft",
legend = c("true relationship",paste0("this instance (seed = ",seed,")")),
lty = c(1,2)
)
}
## Put it in a big data frame -------------------------------------------------
dat <- tibble(
groups = as.factor(groups),
y_obs = y_obs,
x_obs = x_obs,
y_true = rep(y_true,each=n_per_group),
x_true = rep(x_true,each=n_per_group)
) %>%
group_by(groups) %>%
mutate(
y_obs_mean_per_group = mean(y_obs),
x_obs_mean_per_group = mean(x_obs)
) %>%
ungroup()
### Add missing values ----------------------------------------------------
if(miss_type=="random") {
assertthat::assert_that(missingness>=0 && missingness < 1)
mis <- if_else(!rbinom((n_groups*n_per_group),1,missingness),1,NA)
dat <- dat %>%
mutate(x_obs = x_obs*mis)
} else {
is.whole <- function(x, tol = .Machine$double.eps^0.5) abs(x - round(x)) < tol
assertthat::assert_that(is.whole(missingness))
mis <- sample(unique(dat$groups),size = missingness)
dat <- dat %>%
mutate(x_obs = if_else(groups %in% mis,NA,x_obs))
}
attr(dat,"true_intercept") <- y_int_mean
attr(dat,"true_slope") <- x_coef
if(output_type == "full"){
return(dat)
} else {
return(
dat %>%
drop_na() %>%
group_by(groups) %>%
filter(n()>1) %>%
summarise(
y_obs1=mean(y_obs),
x_obs1=mean(x_obs),
n=n(),
sd_y=sd(y_obs),
se_y=sd_y/sqrt(n-1),
sd_x=sd(x_obs),
se_x=sd_x/sqrt(n-1)
) %>%
rename(y_obs = y_obs1, x_obs = x_obs1)
)
}
}
library(tidyverse)
library(brms)
options("brms.backend" = "cmdstanr")
library(marginaleffects)
library(tidybayes)
dlist <- lapply(1:10,\(x) make_data(seed=x,missingness = .3))
# set_rescor(FALSE): Default brms behaviour ----------------------------------
bf_no_rescor <- bf(y_obs ~ mi(x_obs) + (1 | groups)) +
bf(x_obs | mi() ~ 1 + (1 | groups))
bmer_no_rescor <- brm_multiple(
bf_no_rescor,
data = dlist,
prior = c(
prior(student_t(3,0,2.5),class=b,resp=yobs)
),
seed=1L,
chains=4,
cores=4,
iter=2000,
combine = FALSE
)
bmer_no_rescor[[1]]
p1 <- lapply(
bmer_no_rescor,
\(x) plot_predictions(
x,
condition = "x_obs",
re_formula = NA,
resp = "yobs",
draw = F
)
) %>%
bind_rows(.id="Model") %>%
ggplot()+
geom_abline(
intercept = attributes(dlist[[1]])$true_intercept,
slope = attributes(dlist[[1]])$true_slope
)+
geom_lineribbon(
aes(x=x_obs,y=estimate,ymin=conf.low,ymax=conf.high,colour=Model,fill=Model),
alpha=0.4
)+
facet_wrap(vars(Model))+
theme_minimal()+
labs(title="Model with set_rescor(FALSE) [default behaviour]")
# set_rescor(TRUE) --------------------------------------------------
bf_rescor <- bf(y_obs ~ mi(x_obs) + (1 | groups)) +
bf(x_obs | mi() ~ 1 + (1 | groups)) +
set_rescor(TRUE)
bmer_rescor <- brm_multiple(
bf_rescor,
data = dlist,
prior = c(
prior(student_t(3,0,2.5),class=b,resp=yobs)
),
seed=1L,
chains=4,
cores=4,
iter=2000,
combine = FALSE
)
bmer_rescor[[1]]
p2 <- lapply(
bmer_rescor,
\(x) plot_predictions(
x,
condition = "x_obs",
re_formula = NA,
resp = "yobs",
draw = F
)
) %>%
bind_rows(.id="Model") %>%
ggplot()+
geom_abline(
intercept = attributes(dlist[[1]])$true_intercept,
slope = attributes(dlist[[1]])$true_slope
)+
geom_lineribbon(
aes(x=x_obs,y=estimate,ymin=conf.low,ymax=conf.high,colour=Model,fill=Model),
alpha=0.4
)+
facet_wrap(vars(Model))+
theme_minimal()+
labs(title="Model with set_rescor(TRUE) [non-default behaviour]")
patchwork::wrap_plots(p1,p2,guides = "collect")
Output of the code:
bmer_no_rescor[[1]]
Family: MV(gaussian, gaussian)
Links: mu = identity
mu = identity
Formula: y_obs ~ mi(x_obs) + (1 | groups)
x_obs | mi() ~ 1 + (1 | groups)
Data: data[[i]] (Number of observations: 100)
Draws: 4 chains, each with iter = 2000; warmup = 1000; thin = 1;
total post-warmup draws = 4000
Multilevel Hyperparameters:
~groups (Number of levels: 20)
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
sd(yobs_Intercept) 0.63 0.15 0.37 0.96 1.01 660 954
sd(xobs_Intercept) 0.27 0.06 0.18 0.41 1.01 603 1193
Regression Coefficients:
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
yobs_Intercept 5.99 0.24 5.55 6.46 1.01 896 1622
xobs_Intercept 0.51 0.06 0.38 0.63 1.00 847 1444
yobs_mix_obs 0.64 0.38 -0.11 1.39 1.01 808 1090
Further Distributional Parameters:
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
sigma_yobs 0.30 0.02 0.26 0.35 1.00 3104 3079
sigma_xobs 0.11 0.01 0.09 0.14 1.00 1599 2389
bmer_rescor[[1]]
Family: MV(gaussian, gaussian)
Links: mu = identity
mu = identity
Formula: y_obs ~ mi(x_obs) + (1 | groups)
x_obs | mi() ~ 1 + (1 | groups)
Data: data[[i]] (Number of observations: 100)
Draws: 4 chains, each with iter = 2000; warmup = 1000; thin = 1;
total post-warmup draws = 4000
Multilevel Hyperparameters:
~groups (Number of levels: 20)
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
sd(yobs_Intercept) 0.37 0.10 0.20 0.60 1.00 861 1469
sd(xobs_Intercept) 0.31 0.07 0.20 0.46 1.00 1100 1463
Regression Coefficients:
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
yobs_Intercept 5.31 0.20 4.88 5.70 1.00 1583 1690
xobs_Intercept 0.48 0.07 0.33 0.62 1.00 823 1341
yobs_mix_obs 2.06 0.36 1.36 2.79 1.00 1289 1570
Further Distributional Parameters:
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
sigma_yobs 0.37 0.04 0.30 0.47 1.00 1229 1915
sigma_xobs 0.11 0.01 0.09 0.14 1.00 1984 2475
Residual Correlations:
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
rescor(yobs,xobs) -0.57 0.12 -0.76 -0.30 1.00 1380 2501
Edit: in the plot the legend title should really be “Data replicate”.
Could someone explain what’s going on here? Why is the rescor element so important? And is the first model actually not misspecified, but instead somehow appropriate given the simulation? Sorry, I’m about to get on a plane so I don’t have time to write a more considered post.
- Operating System: Mint 22.2
- brms Version: 2.23.0