Here is a simple linear model:
where \sigma,\beta,\gamma,u_n are unobserved and y_n,x_n are observed.
On my earlier question, @spinkney pointed out that this isn’t identified:
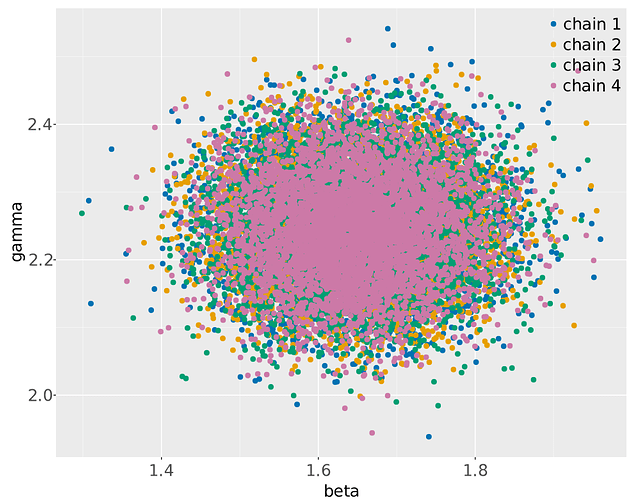
I understand there isn’t enough information to uniquely identify the parameters. I am fine with getting diffuse posteriors. The problem I am facing, however, is that I don’t just get diffuse posteriors, but the chains actually disagree with each other, landing on different modes.
Can I do something differently to get the chains to agree on a diffuse, possibly multi-modal posterior that summarizes the information from the data and likelihood?
data {
int<lower=0> N;
real y[N];
real x[N];
}
parameters {
real beta;
real gamma;
vector[N] u;
real<lower=0> sigma;
}
model {
real x_beta[N];
real u_gamma[N];
to_vector(u) ~ normal(0,1);
beta ~ normal(0, 2);
gamma ~ normal(0, 4);
sigma ~ normal(0,1);
for (n in 1:N) {
x_beta[n] = x[n] * beta;
u_gamma[n] = u[n] * gamma;
y[n] ~ normal(x_beta[n] + u_gamma[n], sigma);
}
}
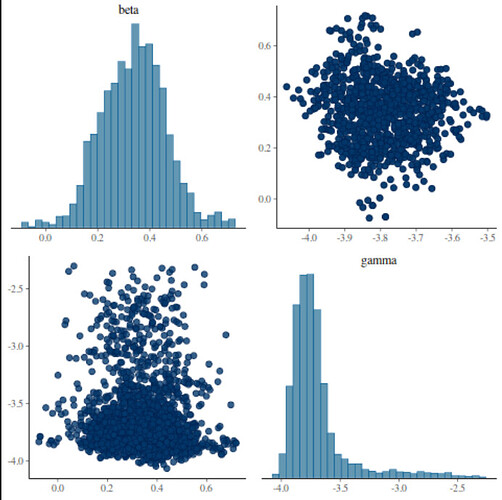
Stansummary shows R_hat exceeds 1 on gamma
Inference for Stan model: train_model
4 chains: each with iter=(1000,1000,1000,1000); warmup=(0,0,0,0); thin=(1,1,1,1); 4000 iterations saved.
Warmup took (5.3, 5.0, 5.0, 5.0) seconds, 20 seconds total
Sampling took (3.8, 3.4, 3.4, 3.4) seconds, 14 seconds total
Mean MCSE StdDev 5% 50% 95% N_Eff N_Eff/s R_hat
lp__ -7.9e+02 2.7e+02 446 -1.8e+03 -6.8e+02 -2.1e+02 2.8 0.20 2.5
accept_stat__ 0.83 1.3e-02 0.17 0.50 0.89 1.00 1.7e+02 1.2e+01 1.0e+00
stepsize__ 0.100 1.7e-02 0.024 0.086 0.086 0.14 2.0e+00 1.4e-01 5.0e+13
treedepth__ 4.5 9.2e-02 0.52 4.0 4.0 5.0 3.3e+01 2.3e+00 1.2e+00
n_leapfrog__ 23 1.6e+00 9.4 15 15 31 3.3e+01 2.3e+00 1.2e+00
divergent__ 0.00 nan 0.00 0.00 0.00 0.00 nan nan nan
energy__ 1294 2.7e+02 447 721 1184 2332 2.8e+00 2.0e-01 2.5e+00
beta 3.4e-01 1.0e-02 0.12 1.5e-01 3.4e-01 5.3e-01 147 10 1.0
gamma -3.7e+00 1.2e-01 0.26 -3.9e+00 -3.8e+00 -3.1e+00 5.0 0.35 1.8
u[1] -6.9e-01 6.4e-03 0.27 -1.1e+00 -7.0e-01 -2.9e-01 1690 120 1.0
u[2] -8.3e-01 9.6e-03 0.28 -1.2e+00 -8.5e-01 -4.1e-01 869 62 1.0
u[3] 1.3e-01 4.0e-03 0.28 -2.7e-01 1.3e-01 5.5e-01 4924 349 1.0
Data generator
Here is the generator:
data {
int<lower=0> N;
}
generated quantities {
real x[N];
real u[N];
for (n in 1:N) {
x[n] = normal_rng(0,1);
u[n] = normal_rng(0,1);
}
real beta = normal_rng(1,1);
real gamma = normal_rng(3,1);
real<lower=0> sigma = normal_rng(0,1);
real x_beta[N];
real u_gamma[N];
real mu[N];
real y[N];
for (n in 1:N) {
x_beta[n] = x[n] * beta;
u_gamma[n] = u[n] * gamma;
mu[n] = x_beta[n] + u_gamma[n];
y[n] = normal_rng(mu[n], sigma);
}
}