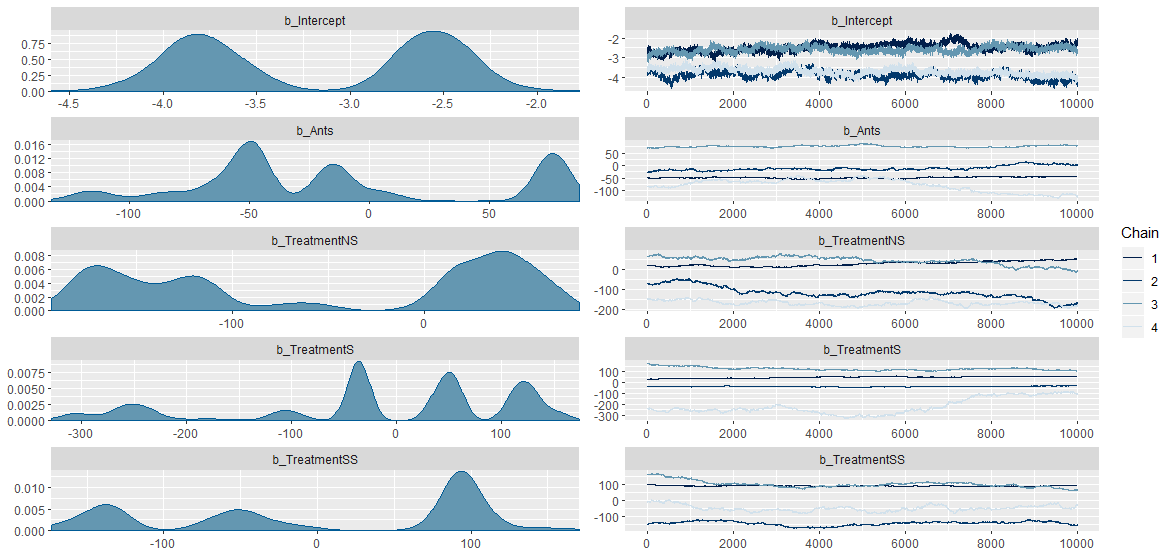
Hi,
I’ve made some models using the brms package and I’ve used the custom_family argument for beta-binomials found in this vignette: add 'beta_binomial2' family · paul-buerkner/custom-brms-families@32c9f2b · GitHub
However, when I try to do post-processing (loo) I get an error for one of my models.
The code I run is as follows:
library(RcmdrMisc)
library(brms)
log_lik_beta_binomial2 <- function(i, draws) {
mu <- draws$dpars$mu[, i]
phi <- draws$dpars$phi
N <- draws$data$trials[i]
y <- draws$data$Y[i]
beta_binomial2_lpmf(y, mu, phi, N)
}
predict_beta_binomial2 <- function(i, draws, ...) {
mu <- draws$dpars$mu[, i]
phi <- draws$dpars$phi
N <- draws$data$trials[i]
beta_binomial2_rng(mu, phi, N)
}
fitted_beta_binomial2 <- function(draws) {
mu <- draws$dpars$mu
trials <- draws$data$trials
trials <- matrix(trials, nrow = nrow(mu), ncol = ncol(mu), byrow = TRUE)
mu * trials
}
# definition of the custom family
beta_binomial2 <- custom_family(
"beta_binomial2",
dpars = c("mu", "phi"),
links = c("logit", "log"),
lb=c(0,0),
ub=c(1,NA),
type = "int", vars = "trials[n]",
log_lik = log_lik_beta_binomial2,
predict = predict_beta_binomial2,
fitted = fitted_beta_binomial2
)
# additionally required Stan code
stan_beta_binomial2 <- "
real beta_binomial2_lpmf(int y, real mu, real phi, int T) {
return beta_binomial_lpmf(y | T, mu * phi, (1 - mu) * phi);
}
real beta_binomial2_rng(real mu, real phi, int T) {
return beta_binomial_rng(T, mu * phi, (1 - mu) * phi);
}
"
added_fun<-stanvar(scode=stan_beta_binomial2,block = "functions")
fit1g=brm(Pest|trials(Trials)~(Ants*Treatment*Plant_species*Insects*Ants_Tree*Ants_Feed*Grass*Day_number)+(1|Block/Tree_ID),data=mydata,family = beta_binomial2, stanvars = added_fun,prior = set_prior("normal(0,10000)",class="b"),chains = 4,iter = 20000)
This code works just fine, however, when I then try to calculate the loo using this code:
expose_functions(fit1g, vectorize = TRUE)
log_lik_beta_binomial2 <- function(i, prep) {
mu <- prep$dpars$mu[, i]
phi <- prep$dpars$phi
trials <- prep$data$vint1[i]
y <- prep$data$Y[i]
beta_binomial2_lpmf(y, mu, phi, trials)
}
fit1g_loo<-loo(fit1g,reloo = TRUE)
I get this error:
Error in (function (y, mu, phi, T, pstream__ = <pointer: 0x00000000019ce540>) :
Exception: beta_binomial_lpmf: First prior sample size parameter is 0, but must be > 0! (in 'unknown file name' at line 5)
Called from: (function (y, mu, phi, T, pstream__ = <pointer: 0x00000000019ce540>)
.Call(<pointer: 0x0000000071282960>, y, mu, phi, T, pstream__))(y = dots[[1L]][[1L]],
mu = dots[[2L]][[6743L]], phi = dots[[3L]][[6743L]], T = dots[[4L]][[1L]])
I’ve searched the forum for similar errors, and see that in cases not using loo but just brm, it might be the init argument that should be changed to something smaller than (-2,2). However, my brm code works fine, so I’m not sure if that is the case here.
Any suggestions for solving the error will be greatly appreciated! :-)
- Operating System: Windows 10
- brms Version: 2.12.0
Best regards,