I’m using a cumulative probit model with Likert data. For this post, I’ve simplified things, in terms of the model structure and the amount of data (I only used ~10% of the data). The model includes varying item thresholds, as well as varying intercepts for items and participants. Model formula as follows:
formula = bf(use | thres(4, gr = item) ~
1 + group +
(1 | item) +
(1 | pid))
Abbreviations: use = dv on a 1-5 likert sale; item = image to be rated; group = between participant categorical factor; pid = participant id.
I tried to attach the reprex model object (model bf.rds), but the forum didn’t allow the file type. Anyway, maybe my question can be answered without it and based on some principles.
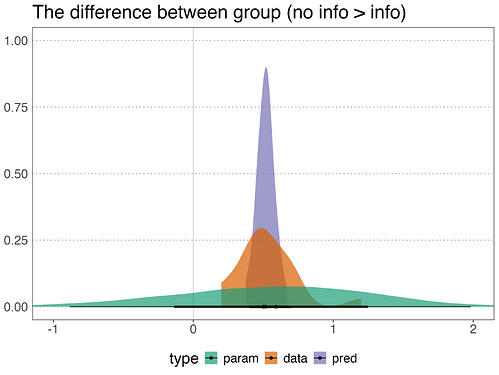
My main question is as follows. It might be fairly basic, as I am inexperienced with these types of model (and most other models, for that matter). In short, the interval estimate for the fixed effects parameter estimate for my main predictor (group: a between participant categorical predictor that indexes the key experimental manipulation) is considerably wider than the interval estimate of the posterior predictions for the same between group difference (see figure below; param = fixed effect of group from the parameter in the model; data = data; pred = posterior predcition via tidybayes add_epred_draws()).
In the past, when I’ve been using other, simpler models, I have not seen such discrepancies between fixed effects and posterior predictions. From what I’ve read (with the possible caveat that I have a bug in my code), my current (mis)understanding goes something like this: the posterior predictions are formed by combining the sum total of parameters in the model, rather than just the fixed effects. And in this model, there are a whole bunch of parameters. For example, in this toy dataset, there are 20 items, each of which has 4 thresholds. And there are 10 pids per group. However, I think I’m still missing an intuition and a justification for why the interval estimates are so discrepant. And this matters, in terms of communicating uncertainty and drawing conclusions, because in one case, the inference would be uncertain as to the sign of the effect (as the interval would cover negative and positive values). In the other case, it would be reasonably certain about the sign (entirely positive). To put it in the crude terms that Reviewer 2 would surely use: “In one case, you show a clear effect of group, whereas on the other case, you do not. What’s going on?”
I would appreciate any help clarifying this matter. And I fully expect I could be confused on multiple levels.
And a more general question about the forum: is there a way to upload a model object to this forum?
FInally, if the data and code help, then I can supply them. But maybe let’s see if the question can be answered first without them.
Thanks.
- Operating System: Mac OS sonoma 14.5
- brms Version: brms 2.21.0