Hi,
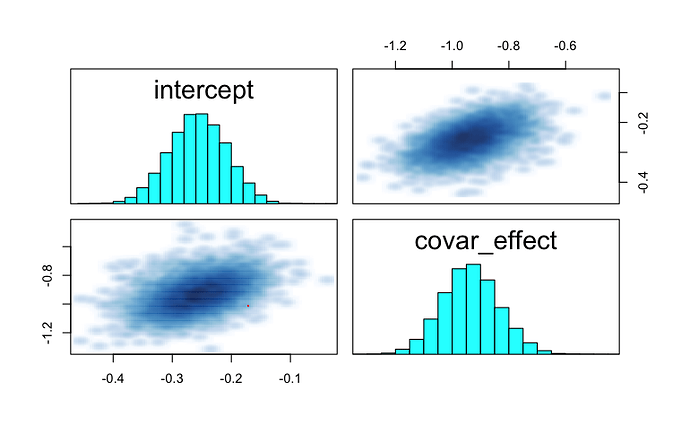
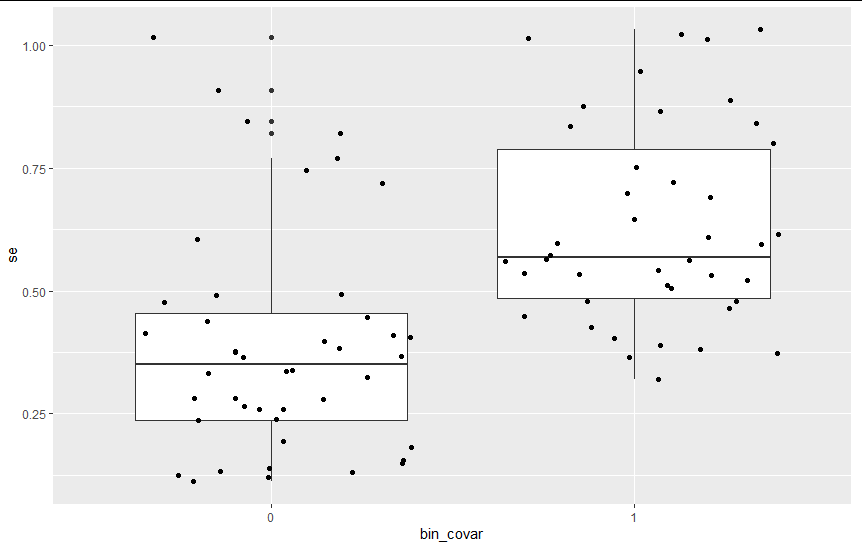
I eventually figured out that the correlation I was getting between my parameters of interest intercept and covar_effect was due to the fact that my binary covariate binary_covar is correlated with the se (higher SE for observations with binary_covar=1).
I’m a bit foggy on why that would lead to correlations in the posterior distributions, and also how bad that is, and also how I could fix it?
Thank you!
my model:
data{
///
int <lower=1> N; // tot observed
int <lower=1> Ngroup; // unique types of
vector[N] expvals;
vector[N] se;
vector[Ngroup] binary_covar;
int<lower=1, upper=Ngroup> group_id[N];
int<lower = 0, upper = 1> run_estimation; // a switch to evaluate the likelihood
}
parameters{
vector[Ngroup] intercept_tilde;
real covar_effect;
real<lower=0> intercept_sd;
real intercept;
}
transformed parameters{
real Xeff_rep[N];
vector[Ngroup] intercept_per_group;
vector[Ngroup] group_mean;
intercept_per_group = intercept_tilde * intercept_sd + intercept;
group_mean = intercept_per_group + (binary_covar - mean(binary_covar) )* covar_effect; //+ ;
for (i in 1:N){
Xeff_rep[i] = group_mean[group_id[i]] ;
}
}
model {
covar_effect ~ normal(0, .5); //
intercept ~ normal(0, .5); //
intercept_sd ~ normal(0, .5); //normal(0, 1);
intercept_tilde ~ std_normal(); //normal(0, 1);
if(run_estimation==1){
expvals ~ normal(Xeff_rep, se); // same as theta (transformed theta-tilde) in 8 schools
}
}
generated quantities{
real eff_rep[N];
for (ix in 1:N){
eff_rep[ix] = normal_rng(group_mean[group_id[ix]], se[ix]);
}
}
Some data:
{"N":[86],"Ngroup":[43],"expvals":[-1.0914,-0.8639,-1.7732,-0.0152,-1.0071,-0.6503,-0.6973,-0.1273,-1.7947,0.3659,-1.1673,-0.5,-1.3925,-0.7708,-0.2552,-0.7064,-1.0336,-0.1527,-0.9017,-0.6918,-1.5845,-0.6171,0.2943,0.1781,0.2469,0.2767,0.1855,0.1515,0.3643,1.1286,0.4952,-0.1314,-0.4427,-0.2831,-1.3268,0.0577,0.4858,-0.3076,0.1341,-0.2223,1.1405,0.0301,0.4403,-1.0552,0.802,-0.2828,-0.9133,-1.3777,-0.1958,-1.1979,-0.7611,-1.2373,-0.9892,-0.3194,-1.6546,-0.5434,-0.4642,-0.5392,-0.6725,-0.3566,0.2213,-0.4259,-0.1281,-0.2893,-0.799,-0.3111,-0.0517,0.4879,0.145,0.1524,0.3558,0.2888,1.1802,0.1139,0.0591,0.4076,0.626,1.3808,-0.2103,0.5548,1.008,0.1728,-0.0534,0.562,0.2475,-1.7548],"se":[0.4788,0.7205,1.0145,0.5652,0.5604,0.4788,0.3721,0.8752,0.8874,0.6975,0.506,0.4642,0.3887,0.5622,0.493,0.5357,0.7515,0.5215,0.3197,0.865,0.609,0.5319,0.377,0.2579,0.1117,0.4388,0.2591,0.2365,0.1942,0.8206,0.1383,0.2816,0.4914,0.3641,0.6041,0.2807,0.4474,0.8455,0.125,0.406,0.339,0.1336,0.7699,0.4028,1.0223,1.0117,0.3652,0.5111,0.6161,0.4261,0.572,1.0315,0.8354,0.6461,0.5972,0.3807,0.6915,0.4103,0.5951,0.8004,0.5424,0.4479,0.9472,0.8415,0.5342,0.3658,0.3318,0.1209,0.4139,0.2654,0.2379,0.182,0.7462,0.1497,0.3245,0.4779,0.3838,0.7193,0.3368,0.374,1.0161,0.1316,0.3972,0.2785,0.1548,0.9091],"binary_covar":[0,0,0,1,1,1,1,0,1,0,1,1,1,0,1,0,1,0,0,0,1,0,0,1,1,0,1,0,0,0,0,1,1,1,0,1,0,0,1,0,1,1,0],"group_id":[4,5,6,7,11,9,12,13,15,17,21,24,25,27,30,32,33,34,36,39,41,42,1,2,3,8,10,14,16,18,19,20,22,23,26,28,29,31,35,37,38,40,43,4,5,6,7,11,9,12,13,15,17,21,24,25,27,30,32,33,34,36,39,41,42,1,2,3,8,10,14,16,18,19,20,22,23,26,28,29,31,35,37,38,40,43],"run_estimation":[1]}
the correlation is 0.49