I’m trying to decide on a sampling distribution for a response variable that is left skewed and slightly bimodal. The response variable is fatty acid concentration of tissues, so observed response values are continuous, and positive.
When I fit a simple intercept model with family = gaussian(link=“identity”), pp_check() indicates negative posterior predictions for the response, which doesn’t make biological sense.
condition_master %>%
filter(lme == "EBS",
!vial_id %in% c("2019-65","2019-67","2019-68","2019-71","2019-66"),
maturity != 1) -> ebs.dat
model_normal <- brm(Total_FA_Conc_WWT ~ 1, family = gaussian(link="identity"), data = ebs.dat)
I tried to remedy this by truncating the response, but I’m not entirely sure I understand the implications of doing this, and the potential trade offs of instead using a model that allows for negative posterior predictions. I also looked at skew normal, gamma and lognormal distributions, and compared how well each model captures the range, mean, min and max of the data. I was unable to run a truncated skew normal model, as it appeared that predictions could not be evaluated properly at the lower end, leading to NA’s.
# fitting a test brms model with a Gaussian likelihood - truncating response at 0, else models predict values < 0
#ie. responses out of bounds are discarded
model_normal <- brm(Total_FA_Conc_WWT | trunc(lb = 0) ~ 1, family = gaussian(link="identity"), data = ebs.dat)
summary(model_normal)
# fitting a test brms model with a skew normal likelihood
model_skew <- brm(Total_FA_Conc_WWT ~ 1, family = skew_normal(), data = ebs.dat)
summary(model_skew)
#If bounded- predictions cannot be evaluated properly at the lower bound, leading to NAs?
#If not bounded, negative posterior predictions for response
# fitting a test brms model with a gamma likelihood
model_gamma <- brm(Total_FA_Conc_WWT ~ 1, family = "gamma", data = ebs.dat)
summary(model_gamma)
# fitting a test brms model with a lognormal likelihood
model_log <- brm(Total_FA_Conc_WWT ~ 1, family = lognormal(), data = ebs.dat)
summary(model_log)
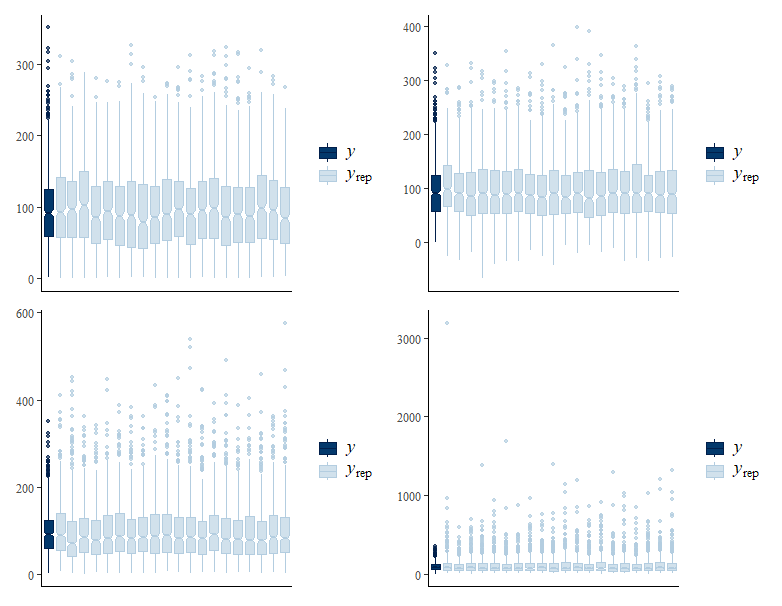
Should have titled, but plots are from top to bottom, L to R: gaussian truncated, skew normal, gamma, and lognormal.
# posterior predictive checking
pp_check(model_normal, ndraws = 1e2) + pp_check(model_skew, ndraws = 1e2) +
pp_check(model_gamma, ndraws = 1e2) + pp_check(model_log, ndraws = 1e2)
#skew normal captures mean and variance best, none of the models picking up apparent bimodality of data
# posterior predictive checking - boxplots
pp_check(model_normal, type = "boxplot", ndraws = 20) + pp_check(model_skew, type = "boxplot", ndraws = 20) +
pp_check(model_gamma, type = "boxplot", ndraws = 20) + pp_check(model_log, type = "boxplot", ndraws = 20)
#and means
pp_check(model_normal, type = "stat", stat = "mean") + pp_check(model_skew, type = "stat", stat = "mean") +
pp_check(model_gamma, type = "stat", stat = "mean") + pp_check(model_log, type = "stat", stat = "mean")
loo_compare(criterion=“waic”) indicates that predictive accuracy is highest for the gaussian truncated model, but I assume that the skew normal model isn’t top solely because I haven’t effectively truncated the response? Log transforming the raw response doesn’t appear to help much either so I’m uncertain which road to take. How “bad” is bad in regards to posterior predictive checks? And should I just be using family=“gaussian” with truncation given the loo results for my model runs that include covariates?
Windows 10
brms_2.19.0