Hi all,
I am new to brms and Bayesian statistics and am having issues with finding a family to fit my outcome distribution.
I have longitudinal data of depressive symptom ratings from 1-10 across 8 weeks. The daily symptom ratings are summed to form a daily total depression score, which is detrended and scaled across the whole sample and time points. This variable will be referred to as TotalDepression_dt_s for convenience. I am trying to model a simple regression with TotalDepresson_dt_s ~ x + y + z + (1 | PlayerID), with x, y, z, being trait/between-person level variables. Essentially, its attempting to model whether trait predictors are associated with trait/average depression.
The issue I’m having is with finding a good family that matches the distribution of TotalDepresson_dt_s.
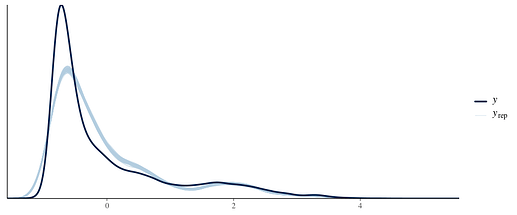
I have tried a gaussian
skew_normal
and exgaussian
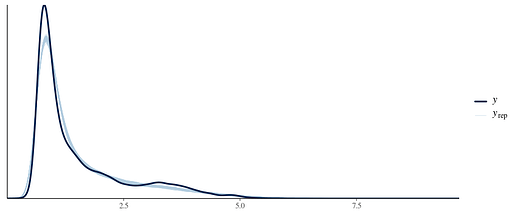
I have also tried shifting the whole distribution to be positive (by adding the min value + 0.001) and fitting lognormal and gamma, which fits similarly well except for the peak. My understanding is that ideally, the family chosen reflects the data generating process-neither of these two families plausibly explains them I think.
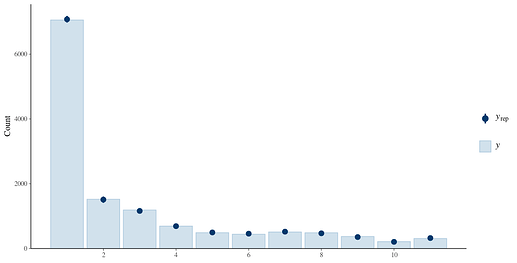
I have considered ordinal families (more specifically for the symptom ratings since they reflect the process the best) but there are too many discrete/unique values. The reason is because the data was detrended for each participant by taking the residuals from Depression ~ AssessmentDay + AssessmentDay^(-0.2) and adding it back to the mean, and then scaled across the whole sample. The variables end up losing discrete ordinal categories and becoming continuous (so this issue extends to the individual symptom ratings as well which share a similar distribution).
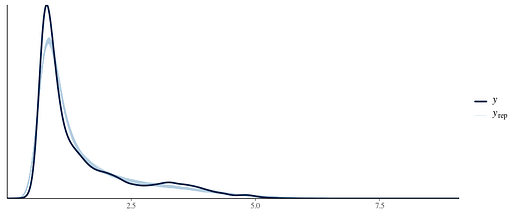
I have considered a gaussian and skew_normal mixture family but it took forever to run. The issue might be my priors but I don’t think a mixed family is the solution to this problem (happy to be convinced otherwise though).
I wonder if there are any other recommendations on what I should/could do?