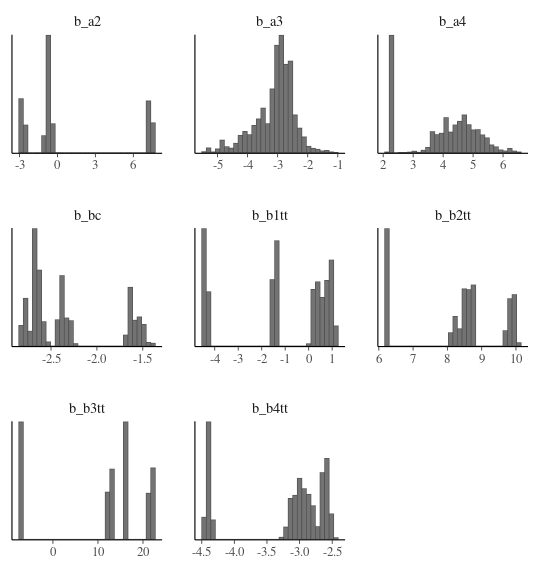
My simplified model is giving slightly more reasonable, albeit still not right, results.
I think one of the reasons for the large sd2’s (which are added to b_a2, b_a3, and b_a4) is that they are counteracting the b_bc, b_b1_tt, b2_tt, b3_tt, and b4_tt (which have sd1’s added to them), which have an exp transformation to give me a LN() rather than N() distribution on them.
I’m running it again with adapt_delta=0.95 (up from 0.90). I’m still confused why it runs with cmdstanr but not cmdstan. Both give a series of messagesException: offset_multiplier_constrain: multiplier[1, 1] is 0, but must be positive finite!, but cmdstanr continues running and stops giving the messages while cmdstan stops with
terminate called without an active exception
[c3208:1143527] *** Process received signal ***
[c3208:1143527] Signal: Aborted (6)
[c3208:1143527] Signal code: (-6)
I’m testing it by running the cmdstan version with startup values from the cmdstanr run (thanks to this thread for the help). I changed my priors a bit and am trying to give it a metric and/or init json input. I can’t seem to get the parameters in the right order for cmdstan though.
./mtc_equity_8 sample num_warmup=1000 num_chains=4 num_threads=32 \
init=test_inits.json \
data file=mtc_data_V3.json \
output file=output_${i}.csv refresh=100 \
adapt delta=0.9 \
random seed=76543
or
./mtc_equity_8 sample num_warmup=1000 num_chains=4 num_threads=32 \
data file=mtc_data_V3.json \
output file=output_${i}.csv refresh=100 \
metric=diag_e metric_file=my_metric1.json \
adapt delta=0.9 \
algorithm=hmc engine=nuts max_depth=10 \
random seed=76543
or
./mtc_equity_8 sample num_warmup=1000 num_chains=4 num_threads=32 \
data file=mtc_data_V3.json \
output file=output_${i}.csv refresh=100 \
adapt delta=0.9 \
random seed=76543