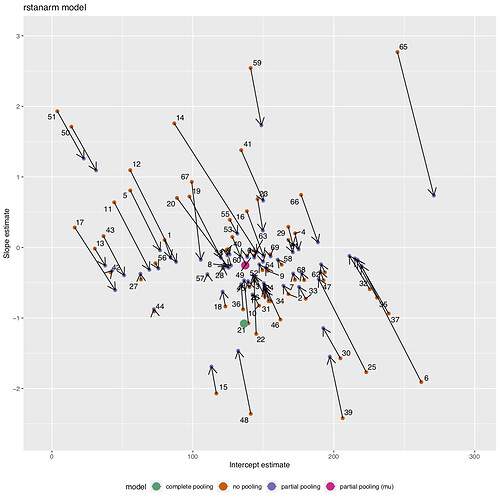
I am building a model with partial pooling, but noticed that some estimates are not being pulled directly to the population-average, and more off to one side. I found this same response to also occur when using test data. Is it possible for models with partial pooling to not always pull directly towards the same point? Does this suggest an issue with my model formatting? Or is it normal, and likely driven by differences in the variance across groups?
For context, I am building a model to analyze long-term data of species life history events (day of flowering, egg laying, reproduction, etc.). Each data point represents the day the event occurred for a given year of observation, but there is high diversity in the species included, type of events, and the length of each observation period can range from 5-30 years. I am primarily interested in seeing how the species slopes are changing over time. To do this, I have been developing a model with random slopes and intercepts and partial pooling for different species. To plot the extent of the partial pooling, I used the vignette found here.
My Stan code is the following:
data {
int<lower=0> N; //No. observations
int<lower=0> Nspp; //No. species
int species[N]; // Grouping by species
vector[N] year;
//response
real ypred[N]; //day of year for nat. history event
}
parameters {
real a[Nspp] ;// intercept for species
real b[Nspp]; // slopes for species
real mu_a; //mean int across sp
real<lower=0> sigma_a; // variation in intercept among species
real mu_b; // mean slope across sp
real<lower=0> sigma_b; //var of slope among sp
real<lower=0> sigma_y; //measurement error
}
transformed parameters{
real mu_y[N]; //individual mean
for(i in 1:N){
mu_y[i]=a[species[i]]+b[species[i]]*year[i];
}
}
model {
a ~ normal(mu_a, sigma_a);
b ~ normal(mu_b, sigma_b);
//Priors
mu_a ~normal(188, 50);
sigma_a ~normal(0,50);
mu_b ~normal(0,10);
sigma_b ~normal(0,10);
sigma_y ~normal(0,10);
ypred ~ normal(mu_y, sigma_y);
}
I would really appreciate any clarification as to whether such observed trends are to be expected given the type of data I am working with or if there are ways I should be adjusting the model itself.
Thanks!