Sorry for bumping this topic up but I recently visited the question. I’ve simulated an extremely simple example and found that this doesn’t seem to work at all.
My main question is why does this not work? It should right?
So, created two data sets that have slightly different beta-values:
# Libraries
library(brms)
library(assortedRFunctions) # devtools::install_github("JAQuent/assortedRFunctions", upgrade = 'never')
cores2use <- 4
samples_for_priors <- 20000
# Generating data 1
n1 <- 100
beta1.0 <- 0
beta1.1 <- 0.2
x <- runif(n1, -1, 1)
y <- beta1.0 + beta1.1*x + rnorm(n1, 0, 1)
data1 <- data.frame(x, y)
# Generating data 2
n2 <- 50
beta2.0 <- 0.2
beta2.1 <- 0.3
x <- runif(n2, -1, 1)
y <- beta2.0 + beta2.1*x + rnorm(n2, 0, 1)
data2 <- data.frame(x, y)
# Combining data sets
data_combined <- rbind(data1, data2)
To test whether chucking everything into one model is the same as first analysing data1 and then data2, I combine data1 & data2 into pooled data frame data_combined.
I then run the first model on data1 and fit a student distrubtion to the posterior distribution for each parameter in this model (Intercept, b_x & sigma). These distributional parameters are used to serve as priors for the model on data2. I also run null models by removing b_x so I can compare various ways of calculating the BFs.
# Sequential analysis
# Data 1
initial_priors <- c(prior(normal(0, 1), class = "Intercept"),
prior(normal(0, 1), class = "b"))
initial_prior_null <- c(prior(normal(0, 1), class = "Intercept"))
m_data1 <- brm(y ~ x,
data = data1,
prior = initial_priors,
chains = 8,
warmup = 2000,
iter = 26000,
cores = cores2use,
save_all_pars = TRUE,
sample_prior = TRUE)
m_data1_null <- brm(y ~ 1,
data = data1,
prior = initial_prior_null,
chains = 8,
warmup = 2000,
iter = 26000,
cores = cores2use,
save_all_pars = TRUE,
sample_prior = TRUE)
# Getting priors for Data 2
# Full model
# Get posterior
postDists <- posterior_samples(m_data1)
samples <- nrow(postDists)
postDists <- postDists[sample(samples, samples_for_priors),]
# Fit model to get distribution
b_Intercept <- brm(b_Intercept ~ 1,
data = postDists,
cores = cores2use,
family = student(link = "identity", link_sigma = "log", link_nu = "logm1"))
# Fit model to get distribution
b_x <- brm(b_x~ 1,
data = postDists,
cores = cores2use,
family = student(link = "identity", link_sigma = "log", link_nu = "logm1"))
# Fit model to get distribution
sigma <- brm(sigma ~ 1,
data = postDists,
cores = cores2use,
family = student(link = "identity", link_sigma = "log", link_nu = "logm1"))
# Fit model to get distribution
priors_data2 <- c(set_prior(priorString_student(b_Intercept),
class = "Intercept"),
set_prior(priorString_student(b_x),
class = "b",
coef = "x"),
set_prior(priorString_student(sigma),
class = 'sigma'))
# Null model
# Get posterior
postDists <- posterior_samples(m_data1_null)
samples <- nrow(postDists)
postDists <- postDists[sample(samples, samples_for_priors),]
# Fit model to get distribution
b_Intercept <- brm(b_Intercept ~ 1,
data = postDists,
cores = cores2use,
family = student(link = "identity", link_sigma = "log", link_nu = "logm1"))
# Fit model to get distribution
sigma <- brm(sigma ~ 1,
data = postDists,
cores = cores2use,
family = student(link = "identity", link_sigma = "log", link_nu = "logm1"))
# Fit model to get distribution
priors_data2_null <- c(set_prior(priorString_student(b_Intercept),
class = "Intercept"),
set_prior(priorString_student(sigma),
class = 'sigma'))
# Run full model for data 2
m_data2 <- brm(y ~ x,
data = data2,
prior = priors_data2,
chains = 8,
warmup = 2000,
iter = 26000,
cores = cores2use,
save_all_pars = TRUE,
sample_prior = TRUE)
# Run null model for data 2
m_data2_null <- brm(y ~ 1,
data = data2,
prior = priors_data2_null,
chains = 8,
warmup = 2000,
iter = 26000,
cores = cores2use,
save_all_pars = TRUE,
sample_prior = TRUE)
I also run a model on the pooled data that:
# Combined analysis
m_data_combined <- brm(y ~ x,
data = data_combined ,
prior = initial_priors,
chains = 8,
warmup = 2000,
iter = 26000,
cores = cores2use,
save_all_pars = TRUE,
sample_prior = TRUE)
m_data_combined_null <- brm(y ~ 1,
data = data_combined ,
prior = initial_prior_null,
chains = 8,
warmup = 2000,
iter = 26000,
cores = cores2use,
save_all_pars = TRUE,
sample_prior = TRUE)
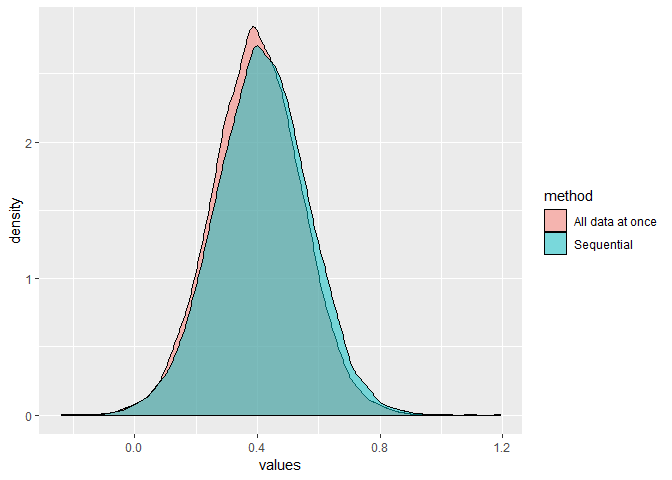
If I compare the posterior distributions for Intercept & b_x from m_data2 and m_data_combined, I see that the distribution seems to be quite different. I used a lot of samples on purpose but it seems very robust.
Look at the difference for the Intercept
and for b_x
This difference is also evidence in the BFs that I calculate in three (two using Savage Dickey ratio & one using bayes_factor()) ways:
# Method: hypothesis()
# Data 1
bf_hypo_1 <- 1/hypothesis(m_data1, 'x = 0')$hypothesis$Evid.Ratio
# Data 2
bf_hypo_2 <- 1/hypothesis(m_data2, 'x = 0')$hypothesis$Evid.Ratio
# Pooled
bf_hypo_pooled <- 1/hypothesis(m_data_combined, 'x = 0')$hypothesis$Evid.Ratio
# Method: Manual with logspline
library(polspline)
# Data 1
priorDist <- prior_samples(m_data1)$b
fit.prior <- logspline(priorDist)
priorDensity <- dlogspline(0, fit.prior)
postDist <- posterior_samples(m_data1)$b_x
fit.posterior <- logspline(postDist)
posterior <- dlogspline(0, fit.posterior)
bf_manual_1 <- priorDensity/posterior
# Data 2
priorDist <- prior_samples(m_data2)$b
fit.prior <- logspline(priorDist)
priorDensity <- dlogspline(0, fit.prior)
postDist <- posterior_samples(m_data2)$b_x
fit.posterior <- logspline(postDist)
posterior <- dlogspline(0, fit.posterior)
bf_manual_2 <- priorDensity/posterior
# Pooled
priorDist <- prior_samples(m_data_combined)$b
fit.prior <- logspline(priorDist)
priorDensity <- dlogspline(0, fit.prior)
postDist <- posterior_samples(m_data_combined)$b_x
fit.posterior <- logspline(postDist)
posterior <- dlogspline(0, fit.posterior)
bf_manual_pooled <- priorDensity/posterior
# Method: bayes_factor()
# Data 1
bf_bayes_factor_1 <- bayes_factor(m_data1, m_data1_null)
# Data 2
bf_bayes_factor_2 <- bayes_factor(m_data2, m_data2_null)
# Pooled
bf_bayes_factor_pooled <- bayes_factor(m_data_combined, m_data_combined_null)
For higher BFs the results acutally often give quite different estimates but for smaller beta they are pretty much in agreement.
BFs from method 1
| Approach |
data1 |
data2 |
combined |
| sequential |
1.97189005763761 |
0.875614188723706 |
1.726615 |
| combined |
|
|
3.027281 |
BFs from method 2
| Approach |
data1 |
data2 |
combined |
| sequential |
2.02109969356174 |
0.828254520749124 |
1.673985 |
| combined |
|
|
3.155711 |
BFs from method 3
| Approach |
data1 |
data2 |
combined |
| sequential |
2.01844391362641 |
0.755140228408023 |
1.524208 |
| combined |
|
|
3.088728 |
Conclusion
I am extremely confused and would love to have some comment on this.
sessionInfo():
R version 3.6.2 (2019-12-12)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows 10 x64 (build 17763)
Matrix products: default
locale:
[1] LC_COLLATE=English_United Kingdom.1252 LC_CTYPE=English_United Kingdom.1252 LC_MONETARY=English_United Kingdom.1252
[4] LC_NUMERIC=C LC_TIME=English_United Kingdom.1252
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] polspline_1.1.19 assortedRFunctions_0.0.1 cowplot_1.0.0 brms_2.12.0 Rcpp_1.0.3
[6] knitr_1.28 ggplot2_3.3.2
loaded via a namespace (and not attached):
[1] Brobdingnag_1.2-6 gtools_3.8.1 StanHeaders_2.21.0-1 threejs_0.3.3 shiny_1.4.0.2
[6] assertthat_0.2.1 highr_0.8 stats4_3.6.2 yaml_2.2.1 backports_1.1.5
[11] pillar_1.4.3 lattice_0.20-38 glue_1.3.2 digest_0.6.25 promises_1.1.0
[16] colorspace_1.4-1 htmltools_0.4.0 httpuv_1.5.2 Matrix_1.2-18 plyr_1.8.6
[21] dygraphs_1.1.1.6 pkgconfig_2.0.3 rstan_2.19.3 purrr_0.3.3 xtable_1.8-4
[26] mvtnorm_1.1-0 scales_1.1.0 processx_3.4.2 later_1.0.0 tibble_2.1.3
[31] farver_2.0.3 bayesplot_1.7.1 DT_0.13 withr_2.1.2 shinyjs_1.1
[36] cli_2.0.2 magrittr_1.5 crayon_1.3.4 mime_0.9 evaluate_0.14
[41] ps_1.3.2 fansi_0.4.1 nlme_3.1-142 xts_0.12-0 pkgbuild_1.0.6
[46] colourpicker_1.0 prettyunits_1.1.1 rsconnect_0.8.16 tools_3.6.2 loo_2.2.0
[51] lifecycle_0.2.0 matrixStats_0.56.0 stringr_1.4.0 munsell_0.5.0 callr_3.4.3
[56] compiler_3.6.2 rlang_0.4.8 grid_3.6.2 ggridges_0.5.2 rstudioapi_0.11
[61] htmlwidgets_1.5.1 crosstalk_1.1.0.1 igraph_1.2.5 miniUI_0.1.1.1 labeling_0.3
[66] base64enc_0.1-3 rmarkdown_2.1 codetools_0.2-16 gtable_0.3.0 inline_0.3.15
[71] abind_1.4-5 markdown_1.1 reshape2_1.4.3 R6_2.4.1 gridExtra_2.3
[76] rstantools_2.0.0 zoo_1.8-7 bridgesampling_1.0-0 dplyr_0.8.5 fastmap_1.0.1
[81] shinystan_2.5.0 shinythemes_1.1.2 stringi_1.4.6 parallel_3.6.2 tidyselect_1.1.0
[86] xfun_0.12 coda_0.19-3