Thank you for the response. I’ve checked the Stan code, but my abilities to read it are quite limited.
It still seems like we’re talking past each other a bit, so let’s simplify the model and the data structure. After making the data, I’ll try to illustrate my basic concern with two simple model. Then I’ll extend the point to variants using ar() and REML.
Data.
Here we make standard bivariate normal data,
\begin{bmatrix} y1 \\ y2 \end{bmatrix} \sim \operatorname{Normal} \left (
\begin{bmatrix} 0 \\ 0 \end{bmatrix},
\begin{bmatrix} 1 & .75 \\ .75 & 1 \end{bmatrix}
\right ),
for which N = 10{,}000. We’ll save the data in both the wide (d_wide) and long (d_long) formats.
# load
library(tidyverse)
library(brms)
library(nlme)
# total sample
n <- 1e4
# poplation correlation
rho <- 0.75
# the variance/covariance matrix
Sigma <-
matrix(c(1, rho, rho, 1),
nrow = 2, byrow = T)
# make a wide data set where you might think of
# y1 and y2 as y values at two time points
set.seed(1)
d_wide <-
mvtnorm::rmvnorm(n = n, sigma = Sigma) %>%
data.frame() %>%
set_names(c("y1", "y2")) %>%
mutate(id = 1:n())
# make a long version of the data with a single criterion called y,
# which is measured at time == 0 and time == 1
d_long <-
d_wide %>%
pivot_longer(-id, values_to = "y") %>%
mutate(time = ifelse(name == "y1", 0, 1))
The data have the sample statistics one would expect.
d_wide %>%
summarise(mean1 = mean(y1),
mean2 = mean(y2),
sd1 = sd(y1),
sd2 = sd(y2),
p = cor(y1, y2))
mean1 mean2 sd1 sd2 p
1 -0.01297871 -0.001211921 1.00779 0.9971651 0.7511297
Simple models.
We’ll start by fitting two simple models. fit1 is a bivariate normal model with no predictors. Thus the \Sigma matrix is reproduced in the parameters. fit2 is captures the correlation by letting y1 predict y2. However, \sigma_2 is now a residual correlation, and thus quite a bit lower (0.66) than it was in fit1.
# bivariate
fit1 <-
brm(data = d_wide,
bf(y1 ~ 1) + bf(y2 ~ 1) + set_rescor(rescor = TRUE))
# pre predicting post
fit2 <-
brm(data = d_wide,
y2 ~ 1 + y1)
This exercize is meant to convey my understanding that when a correlation is expressed within the \Sigma matrix (fit1), I would not expect it to eat into the residual variance the way it would when it is expressed, instead, within the linear model (fit2).
Now we get more to the point.
ar().
Instead of fitting AR(1) models with homogeneous variances, as before, I’m going to switch to heterogeneous variances, which will help showcase the issue.
fit3 <-
brm(data = d_long,
bf(y ~ time + ar(gr = id),
sigma ~ 0 + factor(time)))
This does a great job capturing \rho.
posterior_summary(fit3)["ar[1]", -2] %>% round(digits = 2)
Estimate Q2.5 Q97.5
0.74 0.73 0.76
But look at what happens with the \sigma parameters.
fixef(fit3)[3:4, -2] %>% exp() %>% round(digits = 2)
Estimate Q2.5 Q97.5
sigma_factortime0 1.01 0.99 1.02
sigma_factortime1 0.66 0.65 0.67
Whereas the first is a standard deviation, the second is a residual standard deviation.
ar(cov = TRUE).
Now we explore the results when we set cov = TRUE within the ar() function.
fit4 <-
brm(data = d_long,
bf(y ~ time + ar(gr = id, cov = TRUE),
sigma ~ 0 + factor(time)))
As with the previous model, this one does a great job capturing \rho. For the sake of space, I’ll omit the parameter summary. Of greater interest is what happens to the \sigma parameters.
fixef(fit4)[3:4, -2] %>% exp() %>% round(digits = 2)
Estimate Q2.5 Q97.5
sigma_factortime0 0.67 0.66 0.67
sigma_factortime1 0.66 0.65 0.67
Now they both appear to be in a residual variance metric. Keeping this in mind, consider what happens when we switch to nlme.
nlme with corAR1() and varIdent().
If you look back at the workflow on the IDRE site, Applied Longitudinal Data Analysis, Chapter 7 | R Textbook Examples, you’ll see one can use nlme to fit an AR(1) model with heterogeneous variances using a combination of the corAR1() and varIdent() functions. Here’s an adaptation of their code applied to our simulated data.
fit5 <- gls(data = d_long,
y ~ time,
correlation = corAR1(, form = ~ 1 | id),
weights = varIdent(form = ~1 | time),
method = "REML")
Inspect the results.
print(fit5)
Generalized least squares fit by REML
Model: y ~ time
Data: d_long
Log-restricted-likelihood: -24281.95
Coefficients:
(Intercept) time
-0.01297871 0.01176678
Correlation Structure: AR(1)
Formula: ~1 | id
Parameter estimate(s):
Phi
0.7511297
Variance function:
Structure: Different standard deviations per stratum
Formula: ~1 | time
Parameter estimates:
0 1
1.000000 0.989457
Degrees of freedom: 20000 total; 19998 residual
Residual standard error: 1.00779
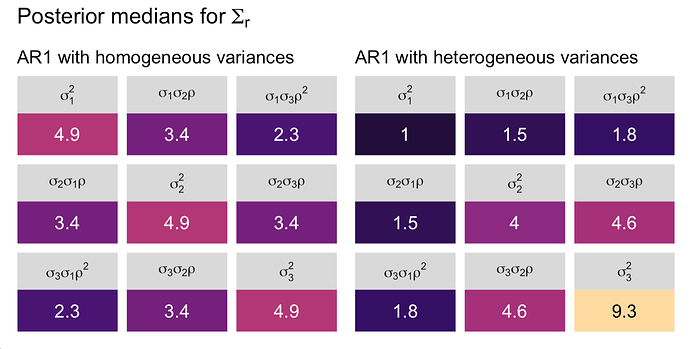
I can’t speak to the equation the nlme package used to compute those \sigma parameters. But it appears, to me, that these residual variances are what I’d expect when (a) the coefficient for time was near zero and the \rho parameter was not eating into the residual variance. This also seems consistent with the models Singer and Willett were discussing in their text. That is, when I fit the model
\begin{align*}
y_{ij} & \sim \operatorname{Normal} (\mu_{ij}, \mathbf r) \\
\mu_{ij} & = \beta_0 + \beta_1 \text{time}_{ij} \\
\mathbf r & = \begin{bmatrix}
\mathbf{\Sigma_r} & \mathbf 0 & \dots & \mathbf 0 \\
\mathbf 0 & \mathbf{\Sigma_r} & \dots & \mathbf 0 \\
\vdots & \vdots & \ddots & \mathbf 0 \\
\mathbf 0 & \mathbf 0 & \mathbf 0 & \mathbf{\Sigma_r}
\end{bmatrix} \\
\mathbf{\Sigma_r} & = \begin{bmatrix}
\sigma_1^2 & \sigma_1 \sigma_2 \rho \\
\sigma_1 \sigma_2 \rho & \sigma_2^2 \end{bmatrix},
\end{align*}
where we already know that \beta_1 = 0 because of how we simulated the data, I would expect the \sigma_1 and \sigma_2 parameters to be very close to 1.
So I suppose this leads me back to my original question: Can one fit this model with brms, whether by using the ar() function or by some other means?