Agreed. Or perhaps there is a more general way to achieve this without any reference to brmsfit objects.
I’ll have to think more about all this. I don’t think it is exactly possible to search by a S3 class.
It would be pleasant if there was a general (non brms specific) approach!
There already is provided that the stanmodel or stanfit object is in the global environment. It shouldn’t recompile unless the model is changed or it is necessary to avoid crashing. Here the problem is finding a stanmodel or stanfit in an essentially arbitrary list.
Right, sorry, I misinterpreted.
I am not sure this thread is related to the question I have, but I have searched in different places (and posted here: https://github.com/quentingronau/bridgesampling/issues/15) without avail. This is the only page that keep popping up for my searches.
I am running brms models and the use bridgesampling to calculate marginal likelihoods of the brms models. I have a loop around all of this. R ‘reliably’ crashes on the second iteration of the loop.
Here is an example that keeps crashing for me (both on a macbook pro or mac pro):
library(tidyverse)
library(magrittr)
library(brms)
library(bridgesampling)
# I've just grabbed this data generator from an old project. It generates a function that can generate
# samples of a by-2 design. The outcome is a binomially distributed response. The function is inelegant
# in a number of ways, but for the present purpose all it needs to do is to generate example data.
data.generator <- function(
estimates = cbind("Intercept" = 0, "Condition" = 0),
n = 2
) {
require(mvtnorm)
require(magrittr)
require(dplyr)
d = data.frame(
Intercept = 1,
Cond = factor(
sort(rep(
c("Control", "Manipulation"),
n / 2)),
levels = c("Manipulation", "Control"))
)
contrasts(d$Cond) = cbind("Condition" = c(1, 0))
d %<>%
mutate(
Condition = case_when(
Cond == "Manipulation" ~ 1,
T ~ 0
)
)
generate.data <- function() {
d$y <- rowSums(d[,colnames(estimates)] * (
matrix(
rep(as.numeric(estimates), n),
byrow = T,
nrow = n
)
))
d$Outcome = sapply(d$y, FUN = function(x) { rbinom(1,1,plogis(x))})
return(d)
}
return(generate.data)
}
# This is the function that fits two brm models and calculates the marginal likelihood for each of
# them using bridgesampling.
fit.replication.models <- function(d.sim2) {
require(brms)
require(bridgesampling)
niter = 8000 # number of iteractions for each chain of brm model
my.priors = c(
prior(normal(0, 10), class = b)
)
my.newpriors.woCondition = my.priors[-1,]
b.rep <- brm(Outcome ~ 1 + Cond,
data = d.sim2, family = bernoulli, iter = niter, warmup = 1000, chains = 4, cores = 4,
prior = my.priors, save_all_pars = T
)
b.rep.woCondition <- brm(Outcome ~ 1,
data = d.sim2, family = bernoulli, iter = niter, warmup = 1000, chains = 4, cores = 4,
prior = my.newpriors.woCondition, save_all_pars = T
)
# Run bridge sampler
bs.rep = bridge_sampler(samples = b.rep, cores = 4, method = "warp3", maxiter = 1000)
bs.rep.woCondition = bridge_sampler(samples = b.rep.woCondition, cores = 4, method = "warp3", maxiter = 1000)
# Store some aspects of the summary of each model, as well as the BF based on the marginal likelihoods obtained
# by the bridge sampler.
simulation <- data.frame(
BF = bf(bs.rep, bs.rep.woCondition)$bf
)
return(simulation)
}
# create a function that generates repeated measures data.
my.data = data.generator(
estimates = cbind("Intercept" = 1, "Condition" = 1),
n = 200
)
# Look at an example data set
my.data()
# run 10 iterations of ta loop that fits brm model and bridgesamples
num.sims = 10
d.sim.orig_rep = plyr::rdply(.n = num.sims,
fit.replication.models(my.data()),
.progress = "text")
Any pointers would be much appreciated.
> version
_
platform x86_64-apple-darwin15.6.0
arch x86_64
os darwin15.6.0
system x86_64, darwin15.6.0
status
major 3
minor 5.2
year 2018
month 12
day 20
svn rev 75870
language R
version.string R version 3.5.2 (2018-12-20)
nickname Eggshell IglooHi tiflo, generally you would want to use the update(brmsfit, newdata = x) function to repeatedly fit a model brmsfit to new datasets x.
Hi Matti, thank you but would I do that even within a loop? I mean if I call, for example,
update(b.rep, data = d.sim2)
in the fit.replication.models function that is repeatedly called in the loop, it should just throw an error because it shouldn’t find the brmsfit object, shouldn’t it? And, fwiw, changing the second model call in fit.replication.models() to
b.rep.woCondition <- update(b.rep, formula. = ~ . - Cond,
iter = niter, warmup = 1000, chains = 4, cores = 4,
prior = my.newpriors.woCondition, save_all_pars = T
)
Still results in R crashing.
And I believe one can’t re-specify priors in update(), correct? That’s relevant because ultimately that is something I will have to do (I’m doing all of this for a replication analysis, which requires me to run the same model on different data sets, using the posterior from one data set as the prior for the other data set).
You can but you need to do a little bit of work upfront by passing your prior hyperparmeters as data via stanvars. See ?stanvars for examples.
Thank you, Paul. That worked in the sense that I now have revised the function to a) only use update() and b) to use stanvars to change the priors. That is, the ‘loop’ now never requires recompiling. However, the function still crashes. I think it might be due to the repeated calls to the bridgesampler. Any idea why that might be happening?
Several people have reported crashing if calling Stan in a loop without small pause after one run stops and the next one is started. Less than one second pause seems to be enough.
Thank you. That seems like an undesirable situation!
Fwiw, adding pauses does not seem to help in this case (thanks to Henrik Singmann for his testing): https://github.com/quentingronau/bridgesampling/issues/15#issuecomment-478356470
I’m currently trying some further debugging and will post the result here.
I have managed to prevent the model from recompiling, but that also does not solve the problem. I’m posting updated code that I’ve tried to simplify and that continues to result in (unfortunately reliable) crashing of R. This would seem to make it impossible to script simulations using brms. Or am I the only one who has this problem?
library(tidyverse)
library(magrittr)
library(brms)
library(bridgesampling)
## ----------------------------------------------------------------------------------
# Functions
## ----------------------------------------------------------------------------------
# Create a data set with a binomially distributed outcome and one binary condition ("Condition").
# Arguments specify the intercept and condition effect, as well as the number of observation n.
data.generator <- function(
estimates = cbind("Intercept" = 0, "Condition" = 0),
n = NA
) {
require(mvtnorm)
require(magrittr)
require(dplyr)
d = data.frame(
Intercept = 1,
Cond = factor(
sort(rep(
c("Control", "Manipulation"),
n / 2)),
levels = c("Manipulation", "Control"))
)
contrasts(d$Cond) = cbind("Condition" = c(1, 0))
d %<>%
mutate(
Condition = case_when(
Cond == "Manipulation" ~ 1,
T ~ 0
)
)
generate.data <- function() {
d$y <- rowSums(d[,colnames(estimates)] * (
matrix(
rep(as.numeric(estimates), n),
byrow = T,
nrow = n
)
))
d$Outcome = sapply(d$y, FUN = function(x) { rbinom(1,1,plogis(x))})
return(d)
}
return(generate.data)
}
fit.replication.models <- function(data1, data2,
model.orig, model.rep.full, model.rep.reduced,
niter = 11000, warmup = 1000, chains = 8, cores = chains, print_priors = T,
n.bs.rep = 1) {
require(brms)
require(bridgesampling)
message(paste0("Sampling with ", niter, " samples per chain for ", chains, " chains (warmup=", warmup, ").\n"))
message(paste0("Using ", cores, " cores.\n"))
cat("Fitting model to original data ...\n")
b.orig <- update(
model.orig,
newdata = data1,
iter = niter, warmup = warmup, chains = chains, cores = cores,
refresh = 0, silent =T
)
p = posterior_samples(b.orig) %>%
dplyr::select(-matches("^(r|lp|cor)_.*")) %<>%
summarise_all(
.funs = c("mean", "sd")
) %>%
gather()
my.rep.stanvars =
stanvar(p[1, "value"], name = "mean_intercept") +
stanvar(p[2, "value"], name = "mean_condition") +
stanvar(p[3, "value"], name = "sd_intercept") +
stanvar(p[4, "value"], name = "sd_condition")
if (print_priors) {
cat("Priors for replication data:\n")
print(p)
}
cat("Fitting model to replication data ...\n")
b.rep <- update(
model.rep.full,
newdata = data2,
stanvars = my.rep.stanvars,
iter = niter, warmup = warmup, chains = chains, cores = cores,
save_all_pars = T,
refresh = 0, silent =T
)
cat("Fitting reduced model to replication data ...\n")
b.rep_reduced <- update(
model.rep.reduced,
newdata = data2,
stanvars = my.rep.stanvars,
iter = niter, warmup = warmup, chains = chains, cores = cores,
save_all_pars = T,
refresh = 0, silent =T
)
simulation = NA
cat("Bridgesampling full model ...\n")
bs.rep = bridge_sampler(samples = b.rep, cores = 8, method = "warp3", maxiter = 1000, repetitions = n.bs.rep)
cat("Bridgesampling reduced model ...\n")
bs.rep_reduced = bridge_sampler(samples = b.rep_reduced, cores = 8, method = "warp3", maxiter = 1000, repetitions = n.bs.rep)
cat("Calculating replication BF and storing information about this analysis ...\n")
s.orig = summary(b.orig)$fixed
s.rep = summary(b.rep)$fixed
simulation <- data.frame(
Int.1.estimate = s.orig[1,1],
Int.1.lower = s.orig[1,3],
Int.1.upper = s.orig[1,4],
Cond.1.estimate = s.orig[2,1],
Cond.1.lower = s.orig[2,3],
Cond.1.upper = s.orig[2,4],
Int.2.estimate = s.rep[1,1],
Int.2.lower = s.rep[1,3],
Int.2.upper = s.rep[1,4],
Cond.2.estimate = s.rep[2,1],
Cond.2.lower = s.rep[2,3],
Cond.2.upper = s.rep[2,4],
BF = bf(bs.rep, bs.rep_reduced)$bf
)
# store simulation results
if (file.exists("simulation.rds"))
simulation = rbind(readRDS("simulation.rds"), simulation)
saveRDS(simulation, "simulation.rds")
# delete all temporary model files
rm(b.orig, b.rep, b.rep_reduced, bs.rep, bs.rep_reduced)
return(simulation)
}
## ----------------------------------------------------------------------------------
# Setting up data generators for original and replication data
## ----------------------------------------------------------------------------------
estimates = cbind("Intercept" = NA, "Condition" = NA)
my.data.orig = data.generator(
estimates = cbind("Intercept" = 1.66, "Condition" = 1),
n = 2000
)
my.data.rep = data.generator(
estimates = cbind("Intercept" = 2.00, "Condition" = 1),
n = 2000
)
## ----------------------------------------------------------------------------------
# Setting up compilation of models
## ----------------------------------------------------------------------------------
formula.full = formula(Outcome ~ 1 + Cond)
formula.reduced = formula(Outcome ~ 1)
# Setting up the values for priors. The specific values here don't matter. We're going to change
# them within the loop anyway.
my.stanvars =
stanvar(0, name = "mean_intercept") +
stanvar(0, name = "mean_condition") +
stanvar(1, name = "sd_intercept") +
stanvar(1, name = "sd_condition")
# Setting up priors for original model. In this case, I'm using a weakly regularizing prior.
# But this detail should not matter.
my.priors = c(prior(normal(0, 10), class = b))
# for reduced replication model
my.reducedpriors = c(prior(normal(mean_intercept, sd_intercept), class = Intercept))
m.fullpriors = my.reducedpriors + prior(normal(mean_condition, sd_condition))
# Compiling and setting up DSO for original model
# These models aren't actually used for anything. I'm just setting them up, so that I can sample from
# them without recompiling later in the loop.
model.orig = brm(
formula = formula.full,
family = bernoulli(),
data = my.data.orig(),
# Using standard uninformative weakly regularizing priors
prior = my.priors,
iter = 2, chains = 1,
save_dso = T, save_model = "m_sim_orig_fullmodel.stan", file = "m_sim_orig_fullmodel"
)
# Compiling and setting up DSO for full model
model.rep.full = brm(
formula = formula.full,
family = bernoulli(),
data = my.data.rep(),
# Setting up priors with stanvars so that they specific values can be manipulated within the loop
prior = m.fullpriors,
stanvars = my.stanvars,
iter = 2, chains = 1,
save_dso = T, save_model = "m_sim_rep_fullmodel.stan", file = "m_sim_rep_fullmodel"
)
# Compiling and setting up DSO for reduced model
model.rep.reduced = brm(
formula = formula.reduced,
family = bernoulli(),
data = my.data.rep(),
# Setting up priors with stanvars so that they specific values can be manipulated within the loop
prior = my.reducedpriors,
stanvars = my.stanvars,
iter = 2, chains = 1,
save_dso = T, save_model = "m_sim_rep_reducedmodel.stan", file = "m_sim_rep_reducedmodel"
)
## ----------------------------------------------------------------------------------
# Running loop
## ----------------------------------------------------------------------------------
num.sims = 50
tictoc::tic()
# requires plyr and tk Set progress bar to "text" if you don't have tcl
d.sim.orig_rep = plyr::rdply(.n = num.sims,
fit.replication.models(
data1 = my.data.orig(),
data2 = my.data.rep(),
model.orig = model.orig,
model.rep.full = model.rep.full,
model.rep.reduced = model.rep.reduced,
niter = 11000, warmup = 1000),
.progress = "tk")
tictoc::toc()You could force avoiding recompilation via recompile = FALSE in update. Perhaps that can help.
Paul, thank you for the suggestion, but the model already doesn’t actually recompile (your suggestions have been successful in this regard). Still, the script crashes in the same way that it did previously during recompilation.
I’ve added recompile = FALSE now and that doesn’t change anything.
This is a problem happening somewhere in rstan or stan itself. Not sure if we have a solution for this problem, yet. @bgoodri do you have any idea?
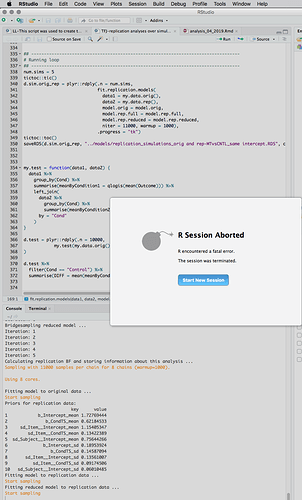
I think so, too. Fwiw, the script always runs through one full iteration of the loop, and then crashes during the second loop – i.e., not the second call to the update() of any model, but the second time update() is called on the same model. To make things even weirder, it seems that it was on the third model for which update() was called for the second time that R crashed (in the screen shot, you can’t see that the script had run through a full iteration [it had], but you can see that the script crashed while sampling for “Fitting reduced model to the replication data”; that’s the third model fit within that function).
Not really. The script runs all the way through for me. Someone who has a Mac will have to try to replicate this problem.